 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_D09G1587 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 567aa MW: 63676.4 Da PI: 4.6324 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 54.5 | 2.9e-17 | 139 | 197 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpse.ng...kr.krfslgkfgtaeeAakaaiaarkkleg 55
++y+GVr++ +g+WvAeIr p + n k r +lg+f a +Aa a+++a+k+++g
Gh_D09G1587 139 CKYRGVRQRI-WGKWVAEIRQPINgNRlasKGnNRLWLGTFSNAIDAALAYDEAAKAMYG 197
79*****998.**********877455997447************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.3E-11 | 139 | 197 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.18E-27 | 139 | 207 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 1.3E-26 | 140 | 206 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 20.784 | 140 | 205 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.37E-17 | 140 | 206 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 1.8E-31 | 140 | 211 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-10 | 141 | 152 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-10 | 187 | 207 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 567 aa Download sequence Send to blast |
MVSPSSIANR RWELKVIPNV SVLLAGQACN MARWVSVVHN ISLEKVLMQG MMEHVVVPMK 60 IKRSSCDTED GVIGSNKKPR ARRNGRESVE DTIEKWKKYN NNQLGLGEED GLKKVSKLPA 120 KGSKKGCMQG KGGPENSRCK YRGVRQRIWG KWVAEIRQPI NGNRLASKGN NRLWLGTFSN 180 AIDAALAYDE AAKAMYGSYA RLNFPDHHSK ESVDNSTTEI GSIESTSTST LDDKRASYET 240 PAVEPVEESK VLLEIKSTDK HDYSQVCTEE EAEDAATDAR ASESTGCNSS KELRIHNSNG 300 INELKDEECQ LRNSCMDSDY YNTQTTLERR GTESEVELSH NIIPRPSDFN FRYNYLDNED 360 QDAGITIFEL EPFKDVKVET PVTRENWEGE LAGSIESGKG YSCFFSEDDN LKTELTDSDI 420 YCKPISKMNA EGSISRDEVD VEPGGFNDFY NYISFDDVYD LKRYEPPDSS CQQQLNTPTS 480 VGGSFNRSTN KLESVHSWSG EAIADVKPFA LIRNDKYGLG GGLEESGNQD ETGFDLDHTM 540 EYLRPDVDLD FLEGIKYTDL WFPERGF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 4e-17 | 141 | 205 | 3 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ghi.15888 | 0.0 | ovule | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 109869454 | 0.0 | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00302 | DAP | Transfer from AT2G40340 | Download |
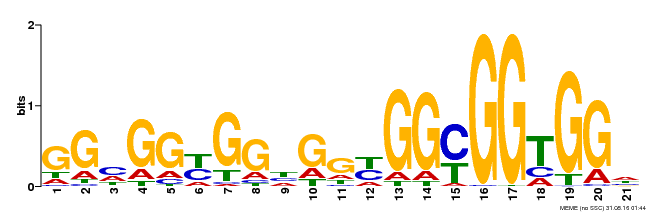
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016671029.1 | 0.0 | PREDICTED: uncharacterized protein LOC107890932 | ||||
| Refseq | XP_016671030.1 | 0.0 | PREDICTED: uncharacterized protein LOC107890932 | ||||
| TrEMBL | A0A1U8HYG4 | 0.0 | A0A1U8HYG4_GOSHI; uncharacterized protein LOC107890932 | ||||
| STRING | EOY07075 | 1e-178 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM28675 | 2 | 3 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38340.1 | 9e-34 | ERF family protein | ||||



