 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.01G183700.1.p | ||||||||
| Common Name | GLYMA_01G183700, LOC100785723 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 368aa MW: 40622.6 Da PI: 7.4775 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 104.7 | 5.3e-33 | 220 | 275 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++W+peLH+rFv+a++qLGG+++AtPk+i+elm+v+gLt+++vkSHLQkYRl+
Glyma.01G183700.1.p 220 KQRRCWSPELHRRFVDALQQLGGAQVATPKQIRELMQVEGLTNDEVKSHLQKYRLH 275
79****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.032 | 217 | 277 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.1E-30 | 218 | 278 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.15E-18 | 218 | 278 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.8E-27 | 220 | 275 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.4E-8 | 222 | 273 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010092 | Biological Process | specification of organ identity | ||||
| GO:0010629 | Biological Process | negative regulation of gene expression | ||||
| GO:0048449 | Biological Process | floral organ formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 368 aa Download sequence Send to blast |
MVVCDFWELS LAQSQRKTPK SNIIMEPSLD LRLGFVPKPL SLFFGDVSGN RDKCDKVVTL 60 DGFVQRLEEE LTKVEAFKRE LPLCILLLND AIARLKEEKV KCSGMQDPPL KTSSGGNENE 120 NSEKKNWMSS AQLWSTQKSK SRNEEDDRSV PANSINGNSC VPEKEGSQVP SFGLMARASE 180 LSHSNSKSVG GDTSSGSSLL RVEVQSQPQP PQHMQQNPRK QRRCWSPELH RRFVDALQQL 240 GGAQVATPKQ IRELMQVEGL TNDEVKSHLQ KYRLHVRRFP VFSIGQVDNG SWMTQDECGD 300 KSKGNMSQSG SPQGPLTPLL LGGAGSAKGL SSPGRNSVDA EDEQSSDCRN WKGGLHHQQL 360 ETDNHSL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 1e-14 | 220 | 273 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 1e-14 | 220 | 273 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 1e-14 | 220 | 273 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 1e-14 | 220 | 273 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.27294 | 0.0 | stem | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00629 | PBM | Transfer from PK14402.1 | Download |
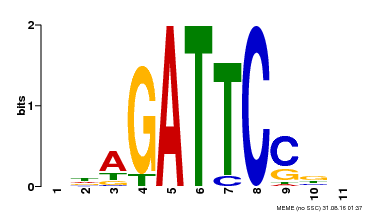
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.01G183700.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015043 | 6e-88 | AP015043.1 Vigna angularis var. angularis DNA, chromosome 10, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003517284.2 | 0.0 | transcription factor HHO5 | ||||
| TrEMBL | I1J945 | 0.0 | I1J945_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA01G39040.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1208 | 34 | 107 | Representative plant | OGRP2563 | 11 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37180.1 | 8e-55 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.01G183700.1.p |
| Entrez Gene | 100785723 |



