 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.01G236300.2.p | ||||||||
| Common Name | GLYMA_01G236300, LOC100804447 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 577aa MW: 62808.5 Da PI: 7.3183 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 51.5 | 2e-16 | 287 | 324 | 3 | 40 |
TCR 3 kkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
+k+CnCkkskClk+YCeCfaag++C e C+C+dC+Nk
Glyma.01G236300.2.p 287 CKRCNCKKSKCLKLYCECFAAGVYCIEPCSCQDCFNKP 324
89**********************************96 PP
| |||||||
| 2 | TCR | 51.1 | 2.7e-16 | 371 | 409 | 1 | 39 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++k+gCnCkks+ClkkYCeC++ g+ Cs +C+Ce+CkN
Glyma.01G236300.2.p 371 RHKRGCNCKKSSCLKKYCECYQGGVGCSISCRCEGCKNA 409
589***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 2.3E-18 | 285 | 326 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 37.11 | 286 | 411 | IPR005172 | CRC domain |
| Pfam | PF03638 | 7.4E-12 | 288 | 323 | IPR005172 | CRC domain |
| SMART | SM01114 | 4.4E-17 | 371 | 412 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 8.6E-12 | 373 | 409 | IPR005172 | CRC domain |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 577 aa Download sequence Send to blast |
MEPKSEGPDC YWDSLIPDAL DMLIFNSPGD AEAFKGLMHK PLASSIRLNE FMSMLPHSTI 60 NNGRKMHIVD SDASGSAPEI EDHCSELMTA TGTNQTQDNF ANVALVTSNS NEKANDQLVS 120 ATHRTIRRRC LDFEMANVQR KNSDENSNTG SSTSESDERN VANEKQLLPA KRNGDLQRGI 180 LQGIGLHLNA LAALKEYKGI QIEKLSSGRQ LSLPSSTSLQ ISTSQEHQNL SLVPVSSERE 240 LDPSDNGVQP AEDCSQPSAY MAGEDFNQNS PKKKRRKSET PGETEGCKRC NCKKSKCLKL 300 YCECFAAGVY CIEPCSCQDC FNKPIHEDTV LQTRKQIESR NPLAFAPKVI RNSDSVPEIG 360 DDPNKTPASA RHKRGCNCKK SSCLKKYCEC YQGGVGCSIS CRCEGCKNAF GRKDGSASVG 420 IETDPEETEA TDKGVTEKAL QKTEIQNTED HPDSATVATP LRLSRPLLPL PFSSKGKPPR 480 SFVTTISGSA LFASQKLGKP NPLWSQAKHF QTVPDEEIPD ILQGDSSPIT CIKTSSPNGK 540 RISSPNCDLG SSPSRRGGRR LILQSIPSFP SLTPHP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 2e-17 | 290 | 408 | 14 | 120 | Protein lin-54 homolog |
| 5fd3_B | 2e-17 | 290 | 408 | 14 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 271 | 275 | KKKRR |
| 2 | 271 | 276 | KKKRRK |
| 3 | 272 | 276 | KKRRK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.41938 | 0.0 | cotyledon| hypocotyl| seed coat| stem | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
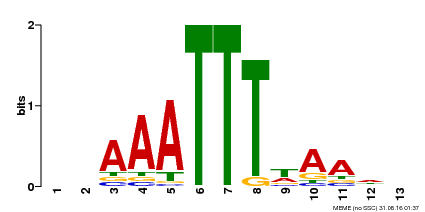
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.01G236300.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003517569.1 | 0.0 | protein tesmin/TSO1-like CXC 2 | ||||
| Refseq | XP_028180593.1 | 0.0 | protein tesmin/TSO1-like CXC 2 | ||||
| TrEMBL | A0A445M7J0 | 0.0 | A0A445M7J0_GLYSO; Protein tesmin/TSO1-like CXC 2 | ||||
| TrEMBL | I1JAR5 | 0.0 | I1JAR5_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA01G44670.1 | 0.0 | (Glycine max) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G14770.1 | 1e-121 | TESMIN/TSO1-like CXC 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.01G236300.2.p |
| Entrez Gene | 100804447 |



