 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.03G003400.2.p | ||||||||
| Common Name | GLYMA_03G003400 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 316aa MW: 35041.3 Da PI: 7.2369 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 56.2 | 7.2e-18 | 246 | 302 | 5 | 61 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklks 61
+r++r++kNRe+A rsR+RK+a+++eLe kv +Le+eN++L+ ++ + + +++ +
Glyma.03G003400.2.p 246 RRQKRMIKNRESAARSRARKQAYTQELEIKVSQLEEENERLRRQNFVWLTHFKSMLQ 302
79****************************************988766666666665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 2.3E-16 | 239 | 289 | No hit | No description |
| SMART | SM00338 | 1.0E-17 | 242 | 306 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.679 | 244 | 289 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.31E-12 | 246 | 297 | No hit | No description |
| Pfam | PF00170 | 8.9E-15 | 246 | 290 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14707 | 1.19E-24 | 246 | 291 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 249 | 264 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 316 aa Download sequence Send to blast |
MGSQSGATQD QEPKTGSLTR QGSLYNLTLD EVQNQLGNLG KPVGSMNLDE LLKSVWTVES 60 GTDAYMHHGG GQVVSAGSSL NPEQGSLTLS GDLSKKTIDE VWRDMQQNKS VGKERQPTLG 120 EMTLEDFLVK AGVSTEPFPN EDGAMAMSGV DSQHNTLQHA HWMQYQLTSV QQQQQPQQQN 180 SVMPGFSGFM AGHVVQQPIP VVLNTVRDAG YSEALPSSLM AALSDSQTAG RKRVASGNVV 240 EKTVERRQKR MIKNRESAAR SRARKQAYTQ ELEIKVSQLE EENERLRRQN FVWLTHFKSM 300 LQESIVMQKR LAWCY* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.26999 | 0.0 | cotyledon| somatic embryo| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in embryo during the latest stages of seed maturation. {ECO:0000269|PubMed:15642716}. | |||||
| Uniprot | TISSUE SPECIFICITY: Predominantly expressed in seeds. {ECO:0000269|PubMed:12376636}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter. Could participate in abscisic acid-regulated gene expression during seed development. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00409 | DAP | Transfer from AT3G56850 | Download |
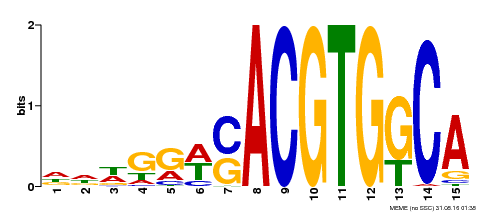
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.03G003400.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015037 | 1e-139 | AP015037.1 Vigna angularis var. angularis DNA, chromosome 4, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003520322.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 2 | ||||
| Refseq | XP_028223942.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 2 | ||||
| Swissprot | Q9LES3 | 1e-88 | AI5L2_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 2 | ||||
| TrEMBL | A0A0R0KPD3 | 0.0 | A0A0R0KPD3_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA03G00580.1 | 0.0 | (Glycine max) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G56850.1 | 1e-79 | ABA-responsive element binding protein 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.03G003400.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



