 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.03G263700.1.p | ||||||||
| Common Name | GLYMA_03G263700, Glyma03g42450.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 346aa MW: 38103.1 Da PI: 4.7758 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 61.1 | 2.5e-19 | 99 | 148 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+y+G+r+++ +g+W+AeIrdp++ r++lg+f taeeAa+a++a +++++g
Glyma.03G263700.1.p 99 QYRGIRQRP-WGKWAAEIRDPRK---GVRVWLGTFNTAEEAARAYDAEARRIRG 148
69*******.**********954...4************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00380 | 1.5E-37 | 99 | 162 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.2E-32 | 99 | 157 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 24.092 | 99 | 156 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.51E-32 | 99 | 158 | No hit | No description |
| SuperFamily | SSF54171 | 6.28E-22 | 99 | 156 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 7.4E-12 | 100 | 148 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.6E-12 | 100 | 111 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.6E-12 | 122 | 138 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0070483 | Biological Process | detection of hypoxia | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 346 aa Download sequence Send to blast |
MCGGAIISDF IPAAAIAGSR RLTADYLWPD LKKRKSDLDV DFEADFRDFK DDSDIDDDDD 60 DHQVKPFAFA ASSRLSTAAK SVAFQGRAEI SANRKRKNQY RGIRQRPWGK WAAEIRDPRK 120 GVRVWLGTFN TAEEAARAYD AEARRIRGKK AKVNFPEAPG TSSVKRSKVN PQENLKTVQP 180 NLGHKFSAGN NHMDLVEQKP LVSQYANMAS FPGSGNGLRS LPSSDDATLY FSSDQGSNSF 240 DYAPEISSML SAPLDCESHF VQNANQQQPN SQNVVSIEDD SAKTLSEELV DIESELKFFQ 300 MPYLEGSWGD TSLESLLSGD TTQDGGNLMN LWCFDDIPSM AGGVF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 2e-20 | 99 | 156 | 5 | 63 | ATERF1 |
| 3gcc_A | 2e-20 | 99 | 156 | 5 | 63 | ATERF1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.31063 | 0.0 | cotyledon| epicotyl| flower| hypocotyl| leaf| meristem| pod| root| stem | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00204 | DAP | Transfer from AT1G53910 | Download |
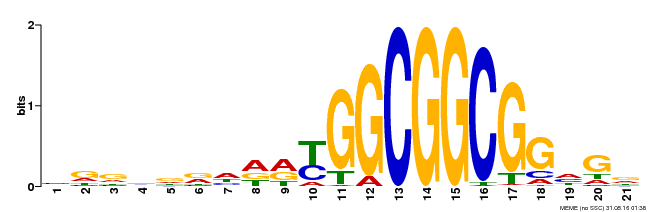
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.03G263700.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT031075 | 0.0 | KT031075.1 Glycine max clone HN_CCL_34 AP2-EREBP transcription factor (Glyma03g42450.1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001238300.2 | 0.0 | EREBP/AP2 transcription factor | ||||
| TrEMBL | A0A0K2CT15 | 0.0 | A0A0K2CT15_SOYBN; AP2-EREBP transcription factor (Fragment) | ||||
| STRING | GLYMA03G42450.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4808 | 32 | 52 | Representative plant | OGRP6 | 16 | 1718 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G53910.3 | 2e-57 | related to AP2 12 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.03G263700.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



