 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.06G199600.1.p | ||||||||
| Common Name | GLYMA_06G199600, LOC102667566 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 403aa MW: 43990.8 Da PI: 8.3789 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 47.3 | 3.7e-15 | 193 | 238 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn+ Er+RRdriN+++ L++l+P+a K +Ka++L ++++Y+k+Lq
Glyma.06G199600.1.p 193 HNQSERKRRDRINQKMKALQRLVPNA-----NKTDKASMLDEVINYLKQLQ 238
9************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.65E-16 | 182 | 242 | No hit | No description |
| SuperFamily | SSF47459 | 1.57E-18 | 186 | 248 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.168 | 188 | 237 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.3E-17 | 193 | 247 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.1E-12 | 193 | 238 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.5E-16 | 194 | 243 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0016607 | Cellular Component | nuclear speck | ||||
| GO:0003690 | Molecular Function | double-stranded DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 403 aa Download sequence Send to blast |
MFPVSMSNYD RISELIWENG QLSIHGLGGL QPTNPTQEKP ISSGAHDTLE SIVQHATFQR 60 YQPSKFTREE GHAPTSNSKN NNSIGAPYYG GEVQGVLSST RKRTWSNANN SMLEECDILS 120 GCASAGATFC RDNDTTMMTW VSLDQSGRSL KTMEEDSACH CGSEIRDNQD DRDSKAEVGQ 180 SNSKRRSRTA AIHNQSERKR RDRINQKMKA LQRLVPNANK TDKASMLDEV INYLKQLQAQ 240 IQMMNMTSMP QMMVPLAMQQ QLQMSMLARM GVAGLGMGMG MNMAAAAQTA GGPIRSLPQF 300 IQPTTTTVCA PPPATAPVFV APSFMMPPSM IQAPPKPQQL PASSASIASI HLPHPYSAAG 360 LTQPINMDIL SNMAALYQQQ ISQNNHQALS SSMPQPHHEQ GR* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 183 | 200 | KRRSRTAAIHNQSERKRR |
| 2 | 196 | 201 | ERKRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00571 | DAP | Transfer from AT5G61270 | Download |
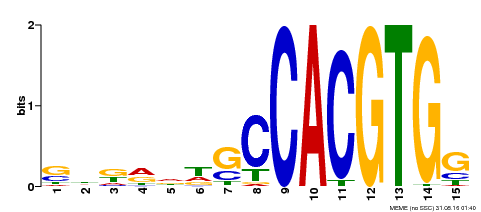
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.06G199600.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006581990.1 | 0.0 | transcription factor PIF7 isoform X1 | ||||
| Refseq | XP_028237308.1 | 0.0 | transcription factor PIF7-like isoform X1 | ||||
| TrEMBL | A0A445KCB0 | 0.0 | A0A445KCB0_GLYSO; Transcription factor PIF7 isoform A | ||||
| TrEMBL | K7KW24 | 0.0 | K7KW24_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA06G21495.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF12999 | 24 | 32 | Representative plant | OGRP258 | 16 | 128 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G61270.1 | 7e-39 | phytochrome-interacting factor7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.06G199600.1.p |
| Entrez Gene | 102667566 |



