 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.07G242000.1.p | ||||||||
| Common Name | GLYMA_07G242000, LOC100793970 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 922aa MW: 104294 Da PI: 7.4451 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 165.3 | 1e-51 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseen 91
+e ++rwl+++ei+a+L n++ +++ ++ + pksg+++L++rk++r+frkDG++wkkkkdgktv+E+he+LKvg+ e +++yYah+++n
Glyma.07G242000.1.p 30 EEaRTRWLRPNEIHAMLCNYKYFTINVKPVNLPKSGTIVLFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGNEERIHVYYAHGQDN 119
4559************************************************************************************** PP
CG-1 92 ptfqrrcywlLeeelekivlvhylevk 118
p+f rrcywlL++++e+ivlvhy+e++
Glyma.07G242000.1.p 120 PNFVRRCYWLLDKSMEHIVLVHYRETQ 146
************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 77.149 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.2E-75 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.1E-45 | 31 | 144 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 8.9E-4 | 366 | 458 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 2.1E-4 | 369 | 455 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 7.0E-15 | 369 | 456 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 3.74E-16 | 554 | 664 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 2.6E-17 | 555 | 668 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 8.24E-19 | 556 | 669 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 9.6E-8 | 562 | 634 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.857 | 572 | 664 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.007 | 605 | 637 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1.1E-5 | 605 | 634 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 940 | 644 | 673 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 2.52E-6 | 752 | 819 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 3.7 | 768 | 790 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.382 | 769 | 798 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.025 | 771 | 789 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0016 | 791 | 813 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.718 | 792 | 816 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0046 | 793 | 813 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 11 | 872 | 894 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.187 | 874 | 902 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045944 | Biological Process | positive regulation of transcription from RNA polymerase II promoter | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001077 | Molecular Function | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 922 aa Download sequence Send to blast |
MANNLAVQLV GSEMHGFHTL QDLDVGSIME EARTRWLRPN EIHAMLCNYK YFTINVKPVN 60 LPKSGTIVLF DRKMLRNFRK DGHNWKKKKD GKTVKEAHEH LKVGNEERIH VYYAHGQDNP 120 NFVRRCYWLL DKSMEHIVLV HYRETQEMQG SPVTPVNSHS SSVSDPPAPW ILSEEIDSGT 180 TTAYTGDMSN NINVKSHELR LHEINTLEWD DLVDTNDHNA STVPNGGTVP YFDQQDQILL 240 NDSFGNVANN LSAEIPSFGN LTQPIAGSNR VPYNFSESVT LQTMDNQANP HEQKNNTVSL 300 SGVDSLDTLV NDRLQSQDSF GMWVNHIMSD SPCSVDDPAL ESPVSSIHEP YSSLVVDSQE 360 SSLPEQVFTI TDVSPTCVSS TEKSKVLVTG FFLKDYMHLS KSNLLCVCGD VSVPAEIVQV 420 GVYRCWVSPH SPGFVNLYLS IDGHKPISQV VNFEYRTPAL HDPAVSMEES DNWDEFRQQM 480 RLAYLLFAKQ LNLDVISSKV SPNRLKEARQ FALKTSFISN SWQYLIKSTE DNQIPFSQAK 540 DALFGITLKN RLKEWLLERI VLGCKTTEYD AHGQSVIHLC AILGYNWAVS LFSWSGLSLD 600 FRDRFGWTAL HWAAYCGREK MVATLLSAGA KPNLVTDPTP QNPGGCTAAD LAYMRGHDGL 660 AAYLSEKSLV QHFNDMSLAG NISGSLETST TDPVNPANLT EDQQNLKDTL TAYRTAAEAA 720 SRIHAAFREH SLKLRTKAVA SSNPEAQARK IVAAMKIQHA FRNHETKKMM AAAARIQCTY 780 RTWKIRKEFL NMRRQAVKIQ AAFRCFQVRK HYRKILWSVG VVEKAVLRWR LKRRGFRGLQ 840 VKTVDAGTGD QDQQSDVEEE FFRTGRKQAE ERVERSVVRV QAMFRSKKAQ EEYRRMKLAL 900 NQAKLEREYE QLLSTEVDMQ L* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.48730 | 0.0 | stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, pollen, top of sepals and siliques. {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
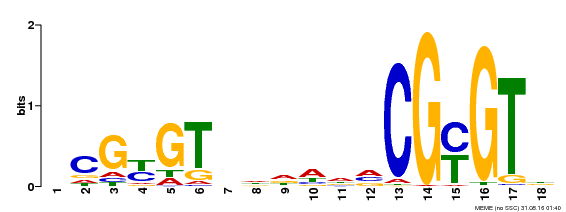
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.07G242000.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015034 | 0.0 | AP015034.1 Vigna angularis var. angularis DNA, chromosome 1, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006584006.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X2 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | K7L3K8 | 0.0 | K7L3K8_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA07G37090.2 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4288 | 34 | 59 | Representative plant | OGRP7351 | 11 | 16 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.07G242000.1.p |
| Entrez Gene | 100793970 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



