 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.09G207300.2.p | ||||||||
| Common Name | GLYMA_09G207300, LOC100798081 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 328aa MW: 36986.1 Da PI: 10.142 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102.4 | 2.8e-32 | 209 | 262 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH+rFv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
Glyma.09G207300.2.p 209 PRMRWTSTLHARFVHAVELLGGHERATPKSVLELMDVKDLTLAHVKSHLQMYRT 262
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.63E-15 | 206 | 262 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-28 | 207 | 262 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 7.1E-24 | 209 | 262 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 5.2E-7 | 210 | 261 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 328 aa Download sequence Send to blast |
MELFPAQPDL SLQISPPNAK PTSSWRRSTE EDMDLGFWKR ALDSRNSIQS MAKQDSCFDL 60 SLSNPKASDN NNNHHSNTTT SNLIHHHHFQ NGATPTNATT TTTNPFQLPF QKNHYFHQQQ 120 QPLFQPQHQS LSQDLGFLRP IRGIPVYQNP PPIPFTQHHH HHHLPLEAST TTPSIISNTN 180 NGSTPFHSQA LMRSRFLSRF PAKRSMRAPR MRWTSTLHAR FVHAVELLGG HERATPKSVL 240 ELMDVKDLTL AHVKSHLQMY RTVKTTDRAA ASSGQSDVYD NGSSGDTSDD LMFDIKSSRR 300 SDLSVKQGRS SVNQDKEYHG LWGNSSR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 1e-16 | 210 | 264 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 1e-16 | 210 | 264 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 1e-16 | 210 | 264 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 1e-16 | 210 | 264 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 1e-16 | 210 | 264 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 1e-16 | 210 | 264 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 1e-16 | 210 | 264 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 1e-16 | 210 | 264 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.32530 | 0.0 | flower| leaf| root | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
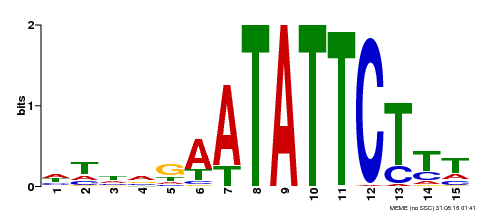
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.09G207300.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015043 | 1e-117 | AP015043.1 Vigna angularis var. angularis DNA, chromosome 10, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006587600.1 | 0.0 | probable transcription factor KAN2 | ||||
| Refseq | XP_028182074.1 | 0.0 | probable transcription factor KAN2 | ||||
| TrEMBL | A0A445J3V0 | 0.0 | A0A445J3V0_GLYSO; Putative transcription factor KAN2 | ||||
| TrEMBL | K7LF36 | 0.0 | K7LF36_SOYBN; Uncharacterized protein | ||||
| TrEMBL | K7LF37 | 0.0 | K7LF37_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA09G34030.2 | 0.0 | (Glycine max) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32240.1 | 2e-64 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.09G207300.2.p |
| Entrez Gene | 100798081 |



