 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.09G217000.1.p | ||||||||
| Common Name | GLYMA_09G217000, LOC100777004 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 655aa MW: 74123.4 Da PI: 6.2385 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 160.3 | 1.2e-49 | 51 | 194 | 1 | 140 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasasp 90
+g+g+k+++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGwvv++DGttyr+ + p+ +++gs+a +s+
Glyma.09G217000.1.p 51 AGKGKKEREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWVVDADGTTYRQ-CPPP---SHMGSFAARSV 136
5899******************************************************************5.4444...69********* PP
DUF822 91 esslq.sslkssalaspvesysaspksssfpspssldsislasa.......asllpvl 140
es+l+ sl++++++ +e++ ++++ ++ +sp+s+ds+ +a+ ++ +p+
Glyma.09G217000.1.p 137 ESQLSgGSLRNCSVKETIENQTSVLRIDECLSPASIDSVVIAERdsktekyTNASPIN 194
*****9***********************************98865555554444444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.2E-50 | 52 | 199 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 1.26E-159 | 217 | 651 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 1.5E-173 | 220 | 650 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 7.3E-88 | 240 | 621 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.7E-58 | 257 | 271 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.7E-58 | 278 | 296 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.7E-58 | 300 | 321 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.7E-58 | 393 | 415 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.7E-58 | 466 | 485 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.7E-58 | 500 | 516 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.7E-58 | 517 | 528 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.7E-58 | 535 | 558 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.7E-58 | 573 | 595 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 655 aa Download sequence Send to blast |
MKSVNDDAST QDLDPQSDHS SDYLAHPQPH PQPRRPRGFA AAAAAATTNS AGKGKKEREK 60 EKERTKLRER HRRAITSRML AGLRQYGNFP LPARADMNDV LAALAREAGW VVDADGTTYR 120 QCPPPSHMGS FAARSVESQL SGGSLRNCSV KETIENQTSV LRIDECLSPA SIDSVVIAER 180 DSKTEKYTNA SPINTVDCLE ADQLMQDIHS GVHENNFTGT PYVPVYVKLP AGIINKFCQL 240 IDPEGIKQEL IHIKSLNVDG VVVDCWWGIV EGWSSQKYVW SGYRELFNII REFKLKLQVV 300 MAFHECGGND SSDALISLPQ WVLDIGKDNQ DIFFTDREGR RNTECLSWGI DKERVLKGRT 360 GIEVYFDMMR SFRTEFDDLF AEGLISAVEV GLGASGELKY PSFSERMGWR YPGIGEFQCY 420 DKYLQHSLRR AAKLRGHSFW ARGPDNAGHY NSMPHETGFF CERGDYDNYY GRFFLHWYSQ 480 TLIDHADNVL SLATLAFEET KITVKVPAVY WWYKTPSHAA ELTAGYHNPT NQDGYSPVFE 540 VLRKHAVTMK FVCLGFHLSS QEANESLIDP EGLSWQVLNS AWDRGLMAAG ENALLCYDRE 600 GYKKLVEIAK PRNDPDRRHF SFFVYQQPSL LQTNVCWSEL DFFVKCMHGE MTDL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-128 | 222 | 612 | 11 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.20912 | 0.0 | leaf| root | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
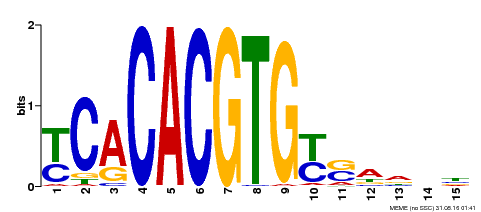
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.09G217000.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC173960 | 0.0 | AC173960.15 Glycine max cultivar Williams 82 clone gmw1-45c9, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003534334.1 | 0.0 | beta-amylase 8 | ||||
| Refseq | XP_028247478.1 | 0.0 | beta-amylase 8-like | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A445J4B9 | 0.0 | A0A445J4B9_GLYSO; Beta-amylase | ||||
| TrEMBL | I1L5A7 | 0.0 | I1L5A7_SOYBN; Beta-amylase | ||||
| STRING | GLYMA09G35070.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 | Representative plant | OGRP9765 | 11 | 12 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.09G217000.1.p |
| Entrez Gene | 100777004 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



