 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.11G131200.1.p | ||||||||
| Common Name | GLYMA_11G131200, Glyma11g13960.1, LOC100789855 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 419aa MW: 45976.7 Da PI: 6.3537 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.6 | 9.4e-13 | 352 | 397 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR +i +++ +L++l+P+ +k + a++L +AveYIk+Lq
Glyma.11G131200.1.p 352 SIAERVRRTKISERMRKLQDLVPNM----DKQTNTADMLDLAVEYIKDLQ 397
689*********************7....688*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 15.186 | 346 | 396 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 9.42E-17 | 349 | 408 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 7.22E-14 | 350 | 401 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 6.1E-16 | 351 | 408 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.1E-9 | 352 | 397 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.4E-14 | 352 | 402 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0045944 | Biological Process | positive regulation of transcription from RNA polymerase II promoter | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0001228 | Molecular Function | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 419 aa Download sequence Send to blast |
MESDLQQHPP MFLDHHQQQM NSGLTRYRSA PSSYFSSIID HEFYEHVFNR PSSPETERML 60 TRFVNSLGGG DADAEDSLAT TQNPPTVVAV KEEVNQQPPD VTSMNNEPLV LQQQQQQQQQ 120 QQQSNNMYNY GSSGTQNFYQ STGRPPLPNQ MKTGHGSSSN LIRHGSSPAG LFSNINIDIT 180 GYAAVVRGMG TMGAAANNTS EEANFSPATR MKNNAPNFSS GLMSSRAEVG NKSNTQNNNA 240 ENEGFAESQG NEFIPAGFPV GPWNDSAIMS DNVTGLKRFR DEDVKPFSGG LNAPESQNET 300 GGQQPSSSAL AHQLSLPNTS AEMAAIEKFL QLSDSVPCKI RAKRGCATHP RSIAERVRRT 360 KISERMRKLQ DLVPNMDKQT NTADMLDLAV EYIKDLQNQV QTLSDNRAKC TCSHKKQQ* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.32036 | 0.0 | hypocotyl| leaf| root| stem | ||||
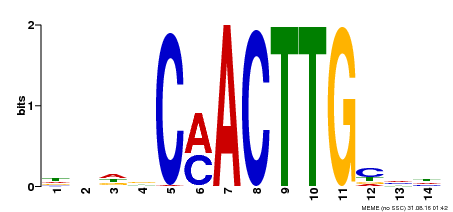
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00195 | DAP | Transfer from AT1G51140 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.11G131200.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT031118 | 0.0 | KT031118.1 Glycine max clone HN_CCL_179 bHLH transcription factor (Glyma11g13960.1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003537954.1 | 0.0 | transcription factor bHLH122 | ||||
| Refseq | XP_006590956.1 | 0.0 | transcription factor bHLH122 | ||||
| Refseq | XP_006590957.1 | 0.0 | transcription factor bHLH122 | ||||
| Refseq | XP_014619508.1 | 0.0 | transcription factor bHLH122 | ||||
| Refseq | XP_028190044.1 | 0.0 | transcription factor bHLH122-like | ||||
| Refseq | XP_028190045.1 | 0.0 | transcription factor bHLH122-like | ||||
| Refseq | XP_028190046.1 | 0.0 | transcription factor bHLH122-like | ||||
| Refseq | XP_028190047.1 | 0.0 | transcription factor bHLH122-like | ||||
| TrEMBL | A0A445I1E2 | 0.0 | A0A445I1E2_GLYSO; Transcription factor bHLH122 isoform A | ||||
| TrEMBL | I1LJT5 | 0.0 | I1LJT5_SOYBN; BHLH transcription factor | ||||
| STRING | GLYMA11G13960.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4185 | 34 | 55 | Representative plant | OGRP511 | 15 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G51140.1 | 8e-83 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.11G131200.1.p |
| Entrez Gene | 100789855 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



