 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.12G184500.2.p | ||||||||
| Common Name | GLYMA_12G184500, LOC100775461 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 488aa MW: 54047.9 Da PI: 7.2442 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 32.2 | 2.3e-10 | 184 | 214 | 4 | 34 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeek 34
k rr+++NReAAr+sR+RKka++++Le
Glyma.12G184500.2.p 184 AKTLRRLAQNREAARKSRLRKKAYVQQLESS 214
6889************************974 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 2.8E-5 | 181 | 246 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.197 | 183 | 227 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.3E-7 | 184 | 216 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14708 | 1.72E-21 | 185 | 235 | No hit | No description |
| SuperFamily | SSF57959 | 1.62E-6 | 185 | 227 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 7.7E-7 | 186 | 227 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 188 | 203 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 2.4E-28 | 266 | 340 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 488 aa Download sequence Send to blast |
MASHRIGELG LSESGPSSHH IPYGVLHGIN TPASSLINQG SAFDFGELEE AIVLHGVKGR 60 NDEGKASLFT ARPAATLEMF PSWPMRFQQT PRVGSKSGGE STDSGLSSKT EPPFEAESPI 120 SKKASSSDHH QAFDQQHLQH RQQLQQEMAS DAPRAASSQN QSAAKSPQEK RKGAGSTSDK 180 PLDAKTLRRL AQNREAARKS RLRKKAYVQQ LESSRLKLNQ IEQELQRARP QGLFVDCGGV 240 GSTVSSGAAM FDMEYARWLE EDHRLMGELR NGLQAPLSDS DMRVMVDGYL SHYDEIFRLK 300 GVAAKSDVFH LINGMWTSQA ERCFLWIGGF RPSDLIMMLI QQLEPLAEQQ IMGMYGLKHS 360 SQQAEEALSQ GLEQLQQSLV DTIAGGPVVD GVQQMVVAMS KLANLEGFVR QADNLRQQTL 420 HQLCRLLTVR QAARCFIVIG EYYGRLRALS SLWASRPRET LINDDNSCQT TTEMQIVQHS 480 QNYFSSF* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.23655 | 0.0 | hypocotyl| root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: During anther development, accumulates in anther primordia during archesporial cell specification and later present in a horseshoe pattern associated with the lateral and adaxial portion of primordia, prior to the emergence of distinct locules. Expressed throughout sporogenic tissue and surrounding cells layers in adaxial and adaxial locules. Localized to the tapetum and middle layers, gradually fading postmeiosis with degeneration of these cell layers. {ECO:0000269|PubMed:20805327}. | |||||
| Uniprot | TISSUE SPECIFICITY: Mostly expressed in stems, inflorescence apex and flowers, and, to a lower extent, in seedlings, leaves and siliques. {ECO:0000269|PubMed:20805327}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Together with TGA10, basic leucine-zipper transcription factor required for anther development, probably via the activation of SPL expression in anthers and via the regulation of genes with functions in early and middle tapetal development (PubMed:20805327). Required for signaling responses to pathogen-associated molecular patterns (PAMPs) such as flg22 that involves chloroplastic reactive oxygen species (ROS) production and subsequent expression of H(2)O(2)-responsive genes (PubMed:27717447). {ECO:0000269|PubMed:20805327, ECO:0000269|PubMed:27717447}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00131 | DAP | Transfer from AT1G08320 | Download |
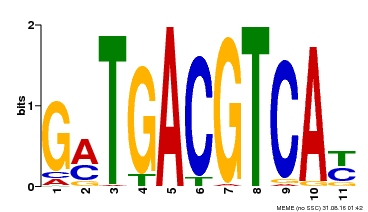
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.12G184500.2.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by flg22 in leaves. {ECO:0000269|PubMed:27717447}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC235234 | 1e-150 | AC235234.1 Glycine max strain Williams 82 clone GM_WBb0028B23, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003539469.1 | 0.0 | transcription factor TGA9 isoform X2 | ||||
| Refseq | XP_028192397.1 | 0.0 | transcription factor TGA9-like isoform X2 | ||||
| Swissprot | Q93XM6 | 0.0 | TGA9_ARATH; Transcription factor TGA9 | ||||
| TrEMBL | A0A0B2R1S2 | 0.0 | A0A0B2R1S2_GLYSO; TGACG-sequence-specific DNA-binding protein TGA-2.1 | ||||
| TrEMBL | K7LVR2 | 0.0 | K7LVR2_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA12G30990.3 | 0.0 | (Glycine max) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08320.3 | 0.0 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.12G184500.2.p |
| Entrez Gene | 100775461 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



