 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.14G030700.2.p | ||||||||
| Common Name | GLYMA_14G030700 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 326aa MW: 37558.4 Da PI: 8.5483 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 160.8 | 5.4e-50 | 36 | 162 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdke 91
pGfrFhPtdeelv +yL++kve+k+l++ e ik++diyk++PwdLp + + +ekewyfF+ r +ky+++ r+nr+t sg+Wkatg dk+
Glyma.14G030700.2.p 36 LPGFRFHPTDEELVGFYLRRKVEKKPLRI-ELIKQIDIYKYDPWDLP-MSSVGEKEWYFFCIRGRKYRNSIRPNRVTGSGFWKATGIDKP 123
69***************************.89***************.4456899*********************************** PP
NAM 92 vlsk..kgelvglkktLvfykgrapkgektdWvmheyrl 128
+++ +e +glkk+Lv+y+g+a kg+ktdW+mhe+rl
Glyma.14G030700.2.p 124 IYCVrePQECIGLKKSLVYYRGSAGKGTKTDWMMHEFRL 162
**98545566***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 3.92E-53 | 33 | 193 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 52.887 | 35 | 194 | IPR003441 | NAC domain |
| Pfam | PF02365 | 4.9E-25 | 37 | 162 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005992 | Biological Process | trehalose biosynthetic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006561 | Biological Process | proline biosynthetic process | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 326 aa Download sequence Send to blast |
MKVPKFSSVA HRLIMDVAKL HNSDDDDDKK EEEVVLPGFR FHPTDEELVG FYLRRKVEKK 60 PLRIELIKQI DIYKYDPWDL PMSSVGEKEW YFFCIRGRKY RNSIRPNRVT GSGFWKATGI 120 DKPIYCVREP QECIGLKKSL VYYRGSAGKG TKTDWMMHEF RLPPNGKTSN NPQANDANDV 180 QEAEVWTLCR IFKRVPTYKK YTPNLKDSAA PITEPNHPTN SSSSKTCSLE SDNCKPYLTF 240 TNSSSPQLIL QNERKPVING HVDVERNHLF LGQLGNVAQA PSFWNHHQNN NVEFANENWD 300 DLTSVVQFAI DPSRVSDHSK EFNRF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 2e-47 | 21 | 200 | 3 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 2e-47 | 21 | 200 | 3 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 2e-47 | 21 | 200 | 3 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 2e-47 | 21 | 200 | 3 | 171 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 2e-47 | 21 | 200 | 3 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 2e-47 | 21 | 200 | 3 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, root caps, cotyledons, tips and margin of young leaves, senescent regions of fully expanded leaves and floral tissues, including old sepals, petals, staments, mature anthers and pollen grains. Not detected in the abscission zone of open flowers, emerging lateral roots and root meristematic zones. {ECO:0000269|PubMed:22345491}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds to the 5'- RRYGCCGT-3' consensus core sequence. Central longevity regulator. Negative regulator of leaf senescence. Modulates cellular H(2)O(2) levels and enhances tolerance to various abiotic stresses through the regulation of DREB2A. {ECO:0000269|PubMed:22345491}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00310 | DAP | Transfer from AT2G43000 | Download |
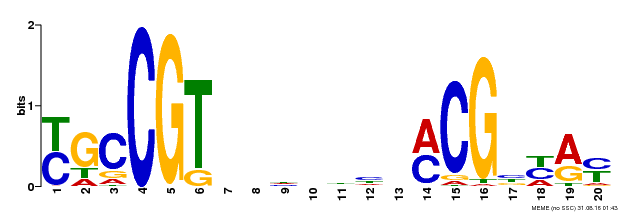
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.14G030700.2.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by H(2)O(2), paraquat, ozone, 3-aminotriazole and salt stress. {ECO:0000269|PubMed:22345491}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT095530 | 0.0 | BT095530.1 Soybean clone JCVI-FLGm-15A17 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028199812.1 | 0.0 | transcription factor JUNGBRUNNEN 1-like isoform X2 | ||||
| Swissprot | Q9SK55 | 1e-92 | NAC42_ARATH; Transcription factor JUNGBRUNNEN 1 | ||||
| TrEMBL | K7M4P1 | 0.0 | K7M4P1_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA14G03441.1 | 0.0 | (Glycine max) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43000.1 | 3e-93 | NAC domain containing protein 42 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.14G030700.2.p |



