 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.14G057900.1.p | ||||||||
| Common Name | GLYMA_14G057900 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 324aa MW: 35573.9 Da PI: 9.0473 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 57.2 | 4.3e-18 | 41 | 90 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+y+GVr+++ +g+WvAeIr+p++ r+r +lg+f tae+Aa+a+++a+ l+g
Glyma.14G057900.1.p 41 RYRGVRQRS-WGKWVAEIREPRK---RTRKWLGTFATAEDAARAYDRAAIILYG 90
59****999.**********954...5**********************98876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 3.06E-20 | 41 | 98 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 2.0E-29 | 41 | 98 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 22.273 | 41 | 98 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.4E-37 | 41 | 104 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 9.81E-21 | 41 | 98 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 5.6E-12 | 41 | 86 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.4E-10 | 42 | 53 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.4E-10 | 64 | 80 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006970 | Biological Process | response to osmotic stress | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0009749 | Biological Process | response to glucose | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010353 | Biological Process | response to trehalose | ||||
| GO:0010449 | Biological Process | root meristem growth | ||||
| GO:0010896 | Biological Process | regulation of triglyceride catabolic process | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:0032880 | Biological Process | regulation of protein localization | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048316 | Biological Process | seed development | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 324 aa Download sequence Send to blast |
KPNHGTHTET TPSETSTVHT NNKISSRKCK GKGGPDNNKF RYRGVRQRSW GKWVAEIREP 60 RKRTRKWLGT FATAEDAARA YDRAAIILYG SRAQLNLQPS GSSSQSSSSR SSSSSTQTLR 120 PLLPRPSGFS VNFPFNTATV PYPYNNSHNY TYSPPVLCPN DNHNNNIVQV HHPHHHRCPE 180 ELVQVQVAHT LSTSYQHNSE SDVGLGVNNN VFVVDGSIRS TSYQRHGFLD SNNNHVHVQV 240 QHGVLSNQQQ QQHQNGVVVE GVNNSPVVGS VASAMDASSV DPDLALVGTV GLGSSSPFWS 300 MANEDDYTVQ IHFAAGDWEA NQR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 7e-19 | 42 | 97 | 3 | 59 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.19347 | 1e-161 | cotyledon| pod| root| somatic embryo | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00674 | SELEX | Transfer from GRMZM2G093595 | Download |
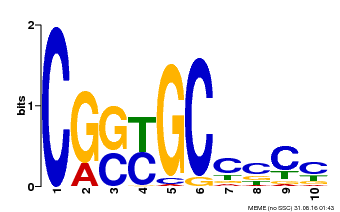
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.14G057900.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC235882 | 0.0 | AC235882.2 Glycine max clone GM_WBc0011M13, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_014622037.1 | 0.0 | LOW QUALITY PROTEIN: ethylene-responsive transcription factor ABI4-like | ||||
| TrEMBL | A0A0R0G9L0 | 0.0 | A0A0R0G9L0_SOYBN; Uncharacterized protein (Fragment) | ||||
| STRING | GLYMA14G06290.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8847 | 29 | 40 | Representative plant | OGRP6 | 16 | 1718 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40220.1 | 2e-36 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.14G057900.1.p |



