 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.14G135400.1.p | ||||||||
| Common Name | GLYMA_14G135400, LOC100127381 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 317aa MW: 34605.4 Da PI: 10.2392 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 98.8 | 3.6e-31 | 239 | 296 | 3 | 59 |
-SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 3 DgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
D+y+WrKYGqK++kgs++pr+YY+C++ gCp++k+ver+ +dp ++++tYegeH+h
Glyma.14G135400.1.p 239 DEYSWRKYGQKPIKGSPYPRGYYKCSTVrGCPARKHVERAPDDPAMLIVTYEGEHRHA 296
9************************9888****************************6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF10533 | 6.8E-17 | 191 | 235 | IPR018872 | Zn-cluster domain |
| Gene3D | G3DSA:2.20.25.80 | 1.5E-32 | 225 | 296 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 30.928 | 232 | 298 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.75E-26 | 235 | 297 | IPR003657 | WRKY domain |
| SMART | SM00774 | 5.1E-37 | 237 | 297 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.1E-26 | 239 | 295 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:1990841 | Molecular Function | promoter-specific chromatin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 317 aa Download sequence Send to blast |
MALELMGFPK LDEQKAIQEA ASEGLKGMEH LIRTLSHQPF HLNTELTDVT VSKFKKLISL 60 LNRTGHARFR RAPVQYSSPP APVHNANTST SSIQLPPPPQ NPNIPAPVQF PSPAPVAVHH 120 APVTLDFTKP HNALLSSNAK SVELEFSKET FSVSSNSSFM SSAITGDGSV SNGKIFLAPP 180 ATSARKPPAF KKRCHEHREH SGDVSGNSKC HCVKRRKNRV KNTVRVPAIS SKIADIPPDE 240 YSWRKYGQKP IKGSPYPRGY YKCSTVRGCP ARKHVERAPD DPAMLIVTYE GEHRHAVQAA 300 MQENAAGVVG LVFEST* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 2e-21 | 225 | 297 | 2 | 73 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gma.27361 | 0.0 | cotyledon| flower| root| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: In young, mature and senescent leaves. {ECO:0000269|PubMed:11722756}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). Regulates rhizobacterium B.cereus AR156-induced systemic resistance (ISR) to P.syringae pv. tomato DC3000, probably by activating the jasmonic acid (JA)- signaling pathway (PubMed:26433201). {ECO:0000250, ECO:0000269|PubMed:26433201}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00464 | DAP | Transfer from AT4G31550 | Download |
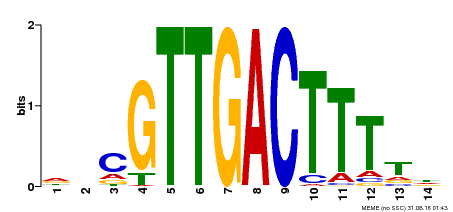
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.14G135400.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Strongly during leaf senescence (PubMed:11722756). Repressed by rhizobacterium B.cereus AR156 in leaves, and to a lower extent, by P.fluorescens WCS417r (PubMed:26433201). {ECO:0000269|PubMed:11722756, ECO:0000269|PubMed:26433201}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU019571 | 0.0 | EU019571.1 Glycine max WRKY31 (WRKY31) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001348293.1 | 0.0 | WRKY transcription factor 31 | ||||
| Refseq | XP_028223284.1 | 0.0 | probable WRKY transcription factor 11 | ||||
| Swissprot | Q9SV15 | 1e-131 | WRK11_ARATH; Probable WRKY transcription factor 11 | ||||
| TrEMBL | A0A445EXZ3 | 0.0 | A0A445EXZ3_GLYSO; Putative WRKY transcription factor 11 | ||||
| TrEMBL | I1M9T5 | 0.0 | I1M9T5_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA14G17730.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1314 | 34 | 98 | Representative plant | OGRP3557 | 11 | 27 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31550.1 | 1e-107 | WRKY DNA-binding protein 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.14G135400.1.p |
| Entrez Gene | 100127381 |



