 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Glyma.15G135100.2.p | ||||||||
| Common Name | GLYMA_15G135100 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 272aa MW: 30016.2 Da PI: 10.1883 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 37.5 | 5.5e-12 | 5 | 56 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqky 47
+++WT+eE++ l+ +v ++G g W+tI + R+ ++k++w+++
Glyma.15G135100.2.p 5 KQKWTAEEEQALKAGVVKHGVGKWRTILKDPEfsgvlYLRSNVDLKDKWRNL 56
79***********************************88***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.947 | 1 | 58 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.72E-16 | 2 | 71 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.7E-7 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-13 | 4 | 56 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.5E-8 | 5 | 56 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 4.87E-19 | 6 | 56 | No hit | No description |
| SuperFamily | SSF46785 | 2.94E-6 | 118 | 156 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 2.3E-6 | 119 | 156 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0006357 | Biological Process | regulation of transcription from RNA polymerase II promoter | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0031627 | Biological Process | telomeric loop formation | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0000784 | Cellular Component | nuclear chromosome, telomeric region | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003691 | Molecular Function | double-stranded telomeric DNA binding | ||||
| GO:0033613 | Molecular Function | activating transcription factor binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0070491 | Molecular Function | repressing transcription factor binding | ||||
| GO:1990841 | Molecular Function | promoter-specific chromatin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 272 aa Download sequence Send to blast |
MGAPKQKWTA EEEQALKAGV VKHGVGKWRT ILKDPEFSGV LYLRSNVDLK DKWRNLSVMA 60 NGWSSREKSR LSVRRVHQVP RQDENSMAIT PVVPSDEEIV DVKPLQVSRD IVHIPGPKRS 120 NLSLDKLIME AITSLKENGG SNKTAIAAFI EVNRKYRIAP IAAYSDRRRN SSMLYLKGRQ 180 KASMKIDRDE TNILTRSQID LELEKIRSMT PQEAAAFAAR AVAEAEAAIA EAEEAAKEAE 240 AAEADAEAAQ AFADATTKSA KGRKSPKRIH T* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats. {ECO:0000269|PubMed:19102728}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00607 | ChIP-seq | Transfer from AT1G49950 | Download |
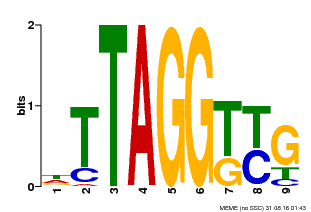
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Glyma.15G135100.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT098318 | 0.0 | BT098318.1 Soybean clone JCVI-FLGm-23G7 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003546290.1 | 1e-157 | telomere repeat-binding factor 1 isoform X2 | ||||
| Refseq | XP_028204581.1 | 1e-157 | telomere repeat-binding factor 1-like isoform X2 | ||||
| Swissprot | Q8VWK4 | 1e-78 | TRB1_ARATH; Telomere repeat-binding factor 1 | ||||
| TrEMBL | I1MG89 | 0.0 | I1MG89_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA15G14320.4 | 1e-156 | (Glycine max) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49950.3 | 1e-79 | telomere repeat binding factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Glyma.15G135100.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



