 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gorai.002G124900.1 | ||||||||
| Common Name | B456_002G124900, LOC105784643 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 337aa MW: 37163.1 Da PI: 10.0077 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 390.2 | 3e-119 | 1 | 336 | 1 | 301 |
GAGA_bind 1 mdddgsre..rnkg.yyepa...aslkenlglqlmssiaerdakirernlalsekkaavaerdmaflqrdkalaernkalverdnkllall 85
mdd g+re r k+ +y+ a +++ +q+m+++aerda+i+ernlalsekkaa+aerdmaflqrd+a+aern+a+ erdn++++l+
Gorai.002G124900.1 1 MDDGGHREngRLKAdQYRTAqgqWLMHQPSMKQIMAIMAERDAAIQERNLALSEKKAAIAERDMAFLQRDAAIAERNNAIAERDNAIANLQ 91
99***999999999999955443345556667*********************************************************** PP
GAGA_bind 86 lvensla....salpvgvqvlsgtksidslqq.lse.pqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkak.kkkk 169
++ensla s++p+g ++++g+k++++ qq +++ p++++ +++ re++ +++lp++ ++ea ++++ k+ ++ak+ ++k+ k+ k
Gorai.002G124900.1 92 YRENSLAsgniSSCPPGFHISRGVKHMQHPQQhVHHlPHISEVPYNSREMHASDVLPVTPGTSEAAKSRQGKRAKEAKVIASNKKAtKPPK 182
*****9999999********************55559********************9988888877777776666665443333224444 PP
GAGA_bind 170 ksekskkkvkkesader.........................skaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCC 235
k ++++++++k + +k+++k dl+ln+v +D+st+ +PvCsCtG+lrqCYkWGnGGWqS+CC
Gorai.002G124900.1 183 KVKQENEDSDKLM---SgkshewkggqdvggagddlnkqlvtTKSDWKGKDLGLNQVVFDDSTMAPPVCSCTGVLRQCYKWGNGGWQSSCC 270
4444444333333...13346799******************************************************************* PP
GAGA_bind 236 tttiSvyPLPvstkrrgaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
tt++S+yPLP+++++r+aRi+grKmS++af+klL +LaaeGydlsnpvDLk+hWAkHGtn+++ti+
Gorai.002G124900.1 271 TTSLSMYPLPAVPNKRHARIGGRKMSGSAFNKLLTRLAAEGYDLSNPVDLKHHWAKHGTNRYITIK 336
*****************************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 5.8E-170 | 1 | 336 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 7.7E-102 | 1 | 336 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 337 aa Download sequence Send to blast |
MDDGGHRENG RLKADQYRTA QGQWLMHQPS MKQIMAIMAE RDAAIQERNL ALSEKKAAIA 60 ERDMAFLQRD AAIAERNNAI AERDNAIANL QYRENSLASG NISSCPPGFH ISRGVKHMQH 120 PQQHVHHLPH ISEVPYNSRE MHASDVLPVT PGTSEAAKSR QGKRAKEAKV IASNKKATKP 180 PKKVKQENED SDKLMSGKSH EWKGGQDVGG AGDDLNKQLV TTKSDWKGKD LGLNQVVFDD 240 STMAPPVCSC TGVLRQCYKW GNGGWQSSCC TTSLSMYPLP AVPNKRHARI GGRKMSGSAF 300 NKLLTRLAAE GYDLSNPVDL KHHWAKHGTN RYITIK* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 174 | 182 | KKATKPPKK |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in seedlings, leaves and pistils. Detected in the base of flowers and tips of carpels, in sepal vasculature, in young rosette, in the lateral and tip of primary roots, and in ovule at the exception of the outer integument. {ECO:0000269|PubMed:14731261, ECO:0000269|PubMed:21435046}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000269|PubMed:14731261}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00540 | DAP | Transfer from AT5G42520 | Download |
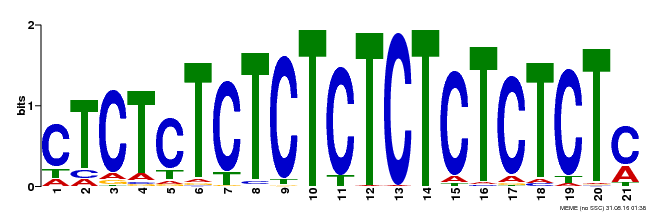
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012465992.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_012465997.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_012466004.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_012466013.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_012466019.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_012466025.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_012466035.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_012466042.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_012466050.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_016707528.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6-like | ||||
| Refseq | XP_016707529.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6-like | ||||
| Refseq | XP_016707530.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6-like | ||||
| Refseq | XP_016707531.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6-like | ||||
| Swissprot | Q8L999 | 1e-149 | BPC6_ARATH; Protein BASIC PENTACYSTEINE6 | ||||
| TrEMBL | A0A0D2Q547 | 0.0 | A0A0D2Q547_GOSRA; Uncharacterized protein | ||||
| TrEMBL | A0A1U8KYL5 | 0.0 | A0A1U8KYL5_GOSHI; protein BASIC PENTACYSTEINE6-like | ||||
| STRING | Gorai.002G124900.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6169 | 26 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42520.1 | 1e-136 | basic pentacysteine 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Gorai.002G124900.1 |
| Entrez Gene | 105784643 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



