 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gorai.002G177000.1 | ||||||||
| Common Name | B456_002G177000, LOC105785690 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 618aa MW: 67860.5 Da PI: 5.34 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 457.1 | 1.1e-139 | 248 | 607 | 1 | 374 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseelaalklfsevsPil 91
lv++L++cAeav++++l+la+al++++ la +++ +m+++a+yf+eALa+r++r + p++ +++ s++l++ f+e++P+l
Gorai.002G177000.1 248 LVHALMACAEAVQQNNLNLAEALVKQIGFLAISQAGAMRKVATYFAEALARRIYR--------FYPQNPLDHSFSDVLHM--HFYETCPYL 328
689****************************************************........44555555557777777..4******** PP
GRAS 92 kfshltaNqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfefnvlv 182
kf+h+taNqaIlea+eg++rvH+iDf+++qG+QWpaL+qaLa R +gpp +R+Tg+g+p++++++ l+e+g++La+fAe+++v+fe++ +v
Gorai.002G177000.1 329 KFAHFTANQAILEAFEGKKRVHVIDFSMNQGMQWPALMQALALRVGGPPAFRLTGIGPPSHDNSDHLQEVGWKLAQFAEKIHVEFEYRGFV 419
******************************************************************************************* PP
GRAS 183 akrledleleeLrvkp..gEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnsesFlerflealeyysalfdsl 271
a++l+dl+ ++L+++p Ea+aVn+v++lh+ll++++++++ v ++vk+++P++v++veqea+hn++ Fl+rf+e+l+yys+lfdsl
Gorai.002G177000.1 420 ANSLADLDASMLDLRPseVEAVAVNSVFELHKLLARPAAIDK----VFSVVKQMKPELVTIVEQEANHNGPVFLDRFTESLHYYSTLFDSL 506
****************8889**************88888888....********************************************* PP
GRAS 272 eaklpreseerikvErellgreivnvvacegaerrerhetlekWrerleeaGFkpvplsekaakqaklllrkvk.sdgyrveeesgslvlg 361
e ++ s++++++++++lg++i+nvvaceg +r+erhe+l +Wr+rl++aGF+pv+l+++a kqa++ll+ ++ +dgy vee++g+l+lg
Gorai.002G177000.1 507 EGSV---SSQDKVMSEVYLGKQICNVVACEGVDRIERHESLTQWRNRLSTAGFSPVHLGSNAFKQASMLLALFAgGDGYGVEENNGCLMLG 594
***7...9999******************************************************************************** PP
GRAS 362 WkdrpLvsvSaWr 374
W++rpL+++SaW+
Gorai.002G177000.1 595 WHNRPLITTSAWK 607
************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01129 | 4.5E-38 | 41 | 114 | No hit | No description |
| Pfam | PF12041 | 1.6E-36 | 41 | 108 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| PROSITE profile | PS50985 | 67.759 | 222 | 586 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 3.8E-137 | 248 | 607 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 618 aa Download sequence Send to blast |
MKRDHNHLQP NPDPSSLLDA PSTGKAKVWD EEAAQQDCGM DELLAVLGYK VKTSDMAEVA 60 QKLEQLEEVM CNVQDDGISH LASETVHYNP SDLSTWLESM LSELNPPSTF DPFGAAGTAA 120 AALDDSFLGP AESSTLTTLD FDNINRKHQK SGQQIFEEAS CSDYDLKAIP GKAIYSQNTQ 180 PQPQTHDSSS SSTPTNVKSE KRFKSTSGPP SPSDIFPPPP PAAASYGIPT DSTRPVVLVD 240 SQENGIRLVH ALMACAEAVQ QNNLNLAEAL VKQIGFLAIS QAGAMRKVAT YFAEALARRI 300 YRFYPQNPLD HSFSDVLHMH FYETCPYLKF AHFTANQAIL EAFEGKKRVH VIDFSMNQGM 360 QWPALMQALA LRVGGPPAFR LTGIGPPSHD NSDHLQEVGW KLAQFAEKIH VEFEYRGFVA 420 NSLADLDASM LDLRPSEVEA VAVNSVFELH KLLARPAAID KVFSVVKQMK PELVTIVEQE 480 ANHNGPVFLD RFTESLHYYS TLFDSLEGSV SSQDKVMSEV YLGKQICNVV ACEGVDRIER 540 HESLTQWRNR LSTAGFSPVH LGSNAFKQAS MLLALFAGGD GYGVEENNGC LMLGWHNRPL 600 ITTSAWKLTK KTTAISQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3g_A | 1e-67 | 243 | 606 | 14 | 378 | Protein SCARECROW |
| 5b3h_A | 1e-67 | 243 | 606 | 13 | 377 | Protein SCARECROW |
| 5b3h_D | 1e-67 | 243 | 606 | 13 | 377 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Gra.3320 | 0.0 | seedling | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that represses transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
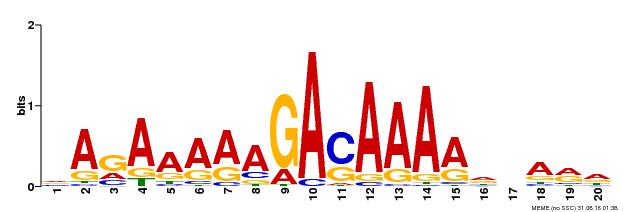
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ643969 | 0.0 | DQ643969.1 Gossypium barbadense DELLA protein GAI (GAI) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012467264.1 | 0.0 | PREDICTED: DELLA protein GAIP-B | ||||
| Swissprot | Q6EI05 | 0.0 | GAIPB_CUCMA; DELLA protein GAIP-B | ||||
| TrEMBL | A0A0D2R838 | 0.0 | A0A0D2R838_GOSRA; Uncharacterized protein | ||||
| STRING | Gorai.002G177000.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM837 | 28 | 121 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01570.1 | 0.0 | GRAS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Gorai.002G177000.1 |
| Entrez Gene | 105785690 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



