 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gorai.002G244200.1 | ||||||||
| Common Name | B456_002G244200, LOC105786382 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 230aa MW: 26896.2 Da PI: 9.1507 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 60.4 | 2.8e-19 | 66 | 119 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+ ++t++ql+ Le+ F+++ +++ +++ +L+++lgL+ rq+ vWFqNrRa++k
Gorai.002G244200.1 66 KKKRLTSDQLDSLEKNFQEEIKLDPDRKMKLSRELGLQPRQIAVWFQNRRARWK 119
56679************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 117.8 | 6.2e-38 | 66 | 156 | 2 | 92 |
HD-ZIP_I/II 2 kkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
kk+rl+++q+ +LE++F+ee kL+p+rK++l+reLglqprq+avWFqnrRAR+k+kqlE+ y++Lk+++da+++e+++L++ev +L+ l+
Gorai.002G244200.1 66 KKKRLTSDQLDSLEKNFQEEIKLDPDRKMKLSRELGLQPRQIAVWFQNRRARWKAKQLERLYDSLKQEFDAISREKQKLQDEVMKLKGILR 156
9*************************************************************************************97665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.2E-18 | 59 | 123 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.281 | 61 | 121 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.7E-16 | 64 | 125 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.1E-16 | 66 | 119 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.64E-15 | 66 | 122 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-20 | 71 | 120 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 4.0E-5 | 92 | 101 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 96 | 119 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 4.0E-5 | 101 | 117 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 5.0E-9 | 121 | 155 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010434 | Biological Process | bract formation | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048510 | Biological Process | regulation of timing of transition from vegetative to reproductive phase | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 230 aa Download sequence Send to blast |
MDWNNTFRPF VSRPEPSLNF LYSNYNYDQY PGMEMKQHQG FMDLGNEMVL PGLNKNSFNN 60 NVNQDKKKRL TSDQLDSLEK NFQEEIKLDP DRKMKLSREL GLQPRQIAVW FQNRRARWKA 120 KQLERLYDSL KQEFDAISRE KQKLQDEVMK LKGILREQVT RNQVSTVYTE ISGEETVEST 180 SIRSSNKPKI AGNNHHPHPV AACNYLFNVD EYNPVSSPYW GTVQLPSYP* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 113 | 121 | RRARWKAKQ |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Widely expressed. {ECO:0000269|PubMed:16055682}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor. {ECO:0000250}. | |||||
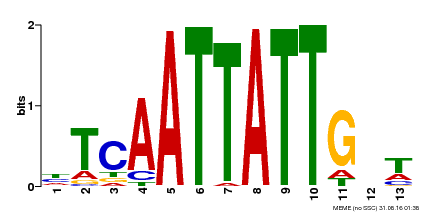
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00486 | DAP | Transfer from AT5G03790 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JX587979 | 1e-69 | JX587979.1 Gossypium hirsutum clone NBRI_GE22268 microsatellite sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012468224.1 | 1e-172 | PREDICTED: homeobox-leucine zipper protein ATHB-22-like | ||||
| Swissprot | Q9LZR0 | 1e-53 | ATB51_ARATH; Putative homeobox-leucine zipper protein ATHB-51 | ||||
| TrEMBL | A0A0D2MFG1 | 1e-170 | A0A0D2MFG1_GOSRA; Uncharacterized protein | ||||
| STRING | Gorai.002G244200.1 | 1e-171 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1660 | 28 | 88 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G03790.1 | 4e-56 | homeobox 51 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Gorai.002G244200.1 |
| Entrez Gene | 105786382 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



