 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gorai.003G085500.1 | ||||||||
| Common Name | B456_003G085500, LOC105788277 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 280aa MW: 31938.7 Da PI: 6.246 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 94.5 | 7.5e-30 | 134 | 191 | 2 | 58 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnh 58
+D ++WrKYGqK++kgs+fprsYYrC+s+ gC ++k+vers +dp+v++itY+ eH h
Gorai.003G085500.1 134 SDVWAWRKYGQKPIKGSPFPRSYYRCSSSkGCLARKQVERSCSDPRVFIITYTAEHCH 191
69*************************998**************************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.9E-30 | 119 | 194 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.891 | 128 | 194 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.31E-25 | 129 | 194 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.5E-35 | 133 | 193 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.5E-24 | 135 | 191 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 280 aa Download sequence Send to blast |
MENWDLQAVV GGNSSNDHLI ANPEFSFGPW SFQQDEDFMS FPEIFETNPK VLDELEQLYK 60 PFYPDLNPFS TQTIITSSIP VPLHVDEPAE KRKKKPSFTV SQSDISASPN PRRFSRKNQQ 120 NRVVEHVTAD DLPSDVWAWR KYGQKPIKGS PFPRSYYRCS SSKGCLARKQ VERSCSDPRV 180 FIITYTAEHC HGHPTRRSSL TGSTRSKPLT AAKSIEAHAE GETVKQERMK MEVELHGEQE 240 GGKILSPDLL LSNDELIRRL EDFDEGFFVD QFPHFSREM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 3e-18 | 121 | 194 | 1 | 74 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in the expression of defense genes in innate immune response of plants. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Activates WRKY 22, SIRK and its own promoters. {ECO:0000269|PubMed:11875555}. | |||||
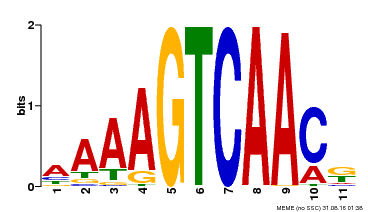
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00448 | DAP | Transfer from AT4G23550 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced 30 minutes after flagellin treatment. {ECO:0000269|PubMed:11875555}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF031074 | 0.0 | KF031074.1 Gossypium hirsutum WRKY transcription factor 12 (WRKY12) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012470538.1 | 0.0 | PREDICTED: probable WRKY transcription factor 29 isoform X1 | ||||
| Swissprot | Q9SUS1 | 6e-55 | WRK29_ARATH; Probable WRKY transcription factor 29 | ||||
| TrEMBL | A0A0D2MGS7 | 0.0 | A0A0D2MGS7_GOSRA; Uncharacterized protein | ||||
| STRING | Gorai.003G085500.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6090 | 25 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G23550.1 | 3e-49 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Gorai.003G085500.1 |
| Entrez Gene | 105788277 |



