 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gorai.007G167100.1 | ||||||||
| Common Name | B456_007G167100, LOC105803425 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 404aa MW: 44163.2 Da PI: 8.2873 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 96.4 | 2e-30 | 93 | 150 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK vkg+ef rsYY+Ct+++C++kk++ers+ d+k+++++Y g+H+h+k
Gorai.007G167100.1 93 QDGYNWRKYGQKLVKGNEFVRSYYKCTHPNCRAKKQLERSH-DGKMIDTVYVGQHDHPK 150
7****************************************.***************85 PP
| |||||||
| 2 | WRKY | 106.7 | 1.1e-33 | 264 | 322 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
++Dgy+WrKYGqK vkg+++prsYYrC+s+gCpvkk+ver+++d k+v++tYeg+H+h+
Gorai.007G167100.1 264 VNDGYRWRKYGQKLVKGNPNPRSYYRCSSPGCPVKKHVERASHDAKLVITTYEGQHDHD 322
58********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 23.548 | 87 | 151 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.5E-26 | 89 | 151 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.01E-24 | 91 | 151 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.1E-32 | 92 | 150 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 7.3E-24 | 93 | 149 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.7E-35 | 250 | 324 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.75E-29 | 257 | 324 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 36.036 | 259 | 324 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.2E-39 | 264 | 323 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.4E-26 | 265 | 321 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009863 | Biological Process | salicylic acid mediated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 404 aa Download sequence Send to blast |
MVPSGECVTD EVGKDKLRSP DAGSHALQHN TDSKIPSSQS DQGGETPLIK SEKDPLSQSE 60 KLGNASSVIT KQTPSLLPGM EGSSPVARER ASQDGYNWRK YGQKLVKGNE FVRSYYKCTH 120 PNCRAKKQLE RSHDGKMIDT VYVGQHDHPK PLNLPLAVGF AVSVVEERPD KSLQIVVKDK 180 SLHSQMPHQI EPRSGSQPLS SAVSDVKGAA SKSNRIQNVA DSDDDHLVSK RRKKENSNAD 240 ASPVEKPTND SRMVIKTFSE VDIVNDGYRW RKYGQKLVKG NPNPRSYYRC SSPGCPVKKH 300 VERASHDAKL VITTYEGQHD HDLPPTRSVT HNTTGVTVHS AAHSDESRTK VEESETVCLD 360 MVVYSGSVAE NKSSEQLNGE LRTKSDFSGT VCVSLIDAPI SGP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 1e-38 | 94 | 325 | 1 | 75 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed to similar levels in root and flower, to a somewhat lower level in stem and to low levels in leaf and siliques. {ECO:0000269|PubMed:8972846}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Binds to a 5'-CGTTGACCGAG-3' consensus core sequence which contains a W box, a frequently occurring elicitor-responsive cis-acting element. {ECO:0000269|PubMed:8972846}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00094 | SELEX | Transfer from AT2G04880 | Download |
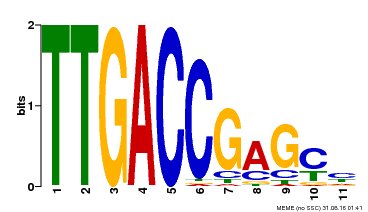
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid (SA). {ECO:0000269|PubMed:17264121}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM438474 | 0.0 | KM438474.1 Gossypium aridum WRKY95 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012491064.1 | 0.0 | PREDICTED: WRKY transcription factor 1-like | ||||
| Swissprot | Q9SI37 | 3e-97 | WRKY1_ARATH; WRKY transcription factor 1 | ||||
| TrEMBL | A0A0D2PAI0 | 0.0 | A0A0D2PAI0_GOSRA; Uncharacterized protein | ||||
| STRING | Gorai.007G167100.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7949 | 26 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G04880.2 | 1e-101 | zinc-dependent activator protein-1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Gorai.007G167100.1 |
| Entrez Gene | 105803425 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



