 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gorai.010G028200.5 | ||||||||
| Common Name | B456_010G028200 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 277aa MW: 30419.1 Da PI: 8.4936 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 32.9 | 1.1e-10 | 117 | 165 | 3 | 55 |
HHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 3 rahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++i +++ L+el+P + +k Ka +L + ++Y++sLq
Gorai.010G028200.5 117 NSHSLAERVRREKISERMKLLQELVPGC----NKITGKAVMLDEIINYVQSLQ 165
58**************************....999*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 2.62E-17 | 112 | 181 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.00E-10 | 112 | 169 | No hit | No description |
| PROSITE profile | PS50888 | 15.586 | 114 | 164 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 3.8E-17 | 115 | 182 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 8.1E-8 | 117 | 165 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.3E-10 | 120 | 170 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 277 aa Download sequence Send to blast |
MNHEGGIERA STNSTQSRDD RQISEEGVVG ASPNRKCRKR VHEANQNADE ELKKDPSGES 60 CDVQKEHDSK KQKTEQNATA NSRGKQVVKQ AKDSSQTGEA PKENYIHVRA RRGQATNSHS 120 LAERVRREKI SERMKLLQEL VPGCNKITGK AVMLDEIINY VQSLQQQVEF LSMKLATVNP 180 ELNIDLERIL SKEILHSRGG NAAVVGFNPG INLSHPFSSG IFPGTISGIP NTNPQFPPFP 240 PQTVLDNELQ NLFQMGFDSS SAMDSLGPIG RLKPGL* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed constitutively in roots, leaves, stems, and flowers. {ECO:0000269|PubMed:12679534}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator involved in cell elongation. Regulates the expression of a subset of genes involved in cell expansion by binding to the G-box motif. Binds to chromatin DNA of the FT gene and promotes its expression, and thus triggers flowering in response to blue light (PubMed:24130508). {ECO:0000269|PubMed:23161888, ECO:0000269|PubMed:24130508}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00135 | DAP | Transfer from AT1G10120 | Download |
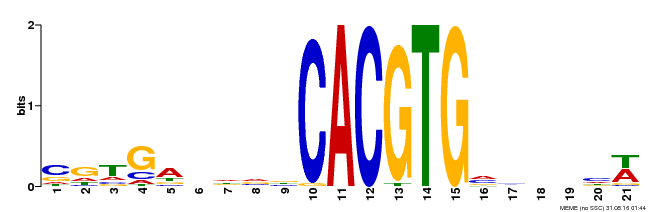
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012451289.1 | 0.0 | PREDICTED: transcription factor bHLH74 | ||||
| Refseq | XP_012451290.1 | 0.0 | PREDICTED: transcription factor bHLH74 | ||||
| Refseq | XP_012451291.1 | 0.0 | PREDICTED: transcription factor bHLH74 | ||||
| Refseq | XP_016726918.1 | 0.0 | PREDICTED: transcription factor bHLH74-like isoform X1 | ||||
| Refseq | XP_016726919.1 | 0.0 | PREDICTED: transcription factor bHLH74-like isoform X1 | ||||
| Swissprot | Q6NKN9 | 7e-89 | BH074_ARATH; Transcription factor bHLH74 | ||||
| TrEMBL | A0A0D2U501 | 0.0 | A0A0D2U501_GOSRA; Uncharacterized protein | ||||
| STRING | Gorai.010G028200.1 | 0.0 | (Gossypium raimondii) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G10120.1 | 2e-85 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Gorai.010G028200.5 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



