 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gorai.013G061100.1 | ||||||||
| Common Name | B456_013G061100, LOC105782485 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1068aa MW: 119815 Da PI: 6.0426 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 183.1 | 3.1e-57 | 20 | 136 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenp 92
l+ ++rwl++ ei++iL n++k+++t+e++trp+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+v+ l+cyYah+een+
Gorai.013G061100.1 20 LEAQHRWLRPAEICEILCNYQKFHITSEPPTRPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSVDKLHCYYAHGEENE 110
5679*************************************************************************************** PP
CG-1 93 tfqrrcywlLeeelekivlvhylevk 118
+fqrr+yw+Le +l +iv+vhylevk
Gorai.013G061100.1 111 NFQRRSYWMLEPDLMHIVFVHYLEVK 136
***********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 86.265 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.3E-84 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 5.7E-51 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 3.9E-6 | 479 | 580 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 2.0E-6 | 494 | 573 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 1.15E-16 | 494 | 580 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF12796 | 2.4E-6 | 675 | 751 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.39E-13 | 676 | 785 | No hit | No description |
| PROSITE profile | PS50297 | 18.351 | 676 | 787 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.4E-17 | 679 | 790 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 3.42E-16 | 679 | 787 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.885 | 726 | 758 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.67E-7 | 886 | 937 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.5 | 886 | 908 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.858 | 887 | 916 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0016 | 888 | 907 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 909 | 931 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.395 | 910 | 934 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.7E-4 | 912 | 931 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1068 aa Download sequence Send to blast |
MADRASYSLA PRLDIEQILL EAQHRWLRPA EICEILCNYQ KFHITSEPPT RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKVGSVDKLH CYYAHGEENE NFQRRSYWML 120 EPDLMHIVFV HYLEVKGSRT IGGIRENSDL SNSQTSSLLT SSNSVTHTKE PSAHANSACS 180 ITSLTSLYED ADSEDSCQAS SRVRTSPQIG NATVMDKLDP GFLNHYSPHP IQGQSSIPGV 240 TEVSHLHGDR TGDTNYGSCI SEAQRTLGLT SWEQGLEPYV PVYADAFSNA SLTSTQPDTI 300 SISLQQETMM KGKLLAVESA GGEFGNPLPT QPHWQTPLAD NALELPKWSM DPSSNFDLPF 360 DSKLFEQNAH EFQNTLEEFS GHGVFNDQPL HKNLQMQIMN ADSHSVMRTY PDNDMHLDGN 420 VNYALSLKKS LLDGEESLKK VDSFSRWVTK ELGEVDNLQM QSSSGIAWST VECGNVSDDA 480 SLSPSLSHDQ LFSIVDFSPK WAYIDLETEV LIIGTYLRSQ EQVAKYNWSC MFGEVEVSAE 540 VIADGILSCY APPHSVGQVP FYVTCSNRLA CSEVREFDYR AGFTKDINIL DIYDIASREM 600 LMRFERLLSL KSSNYPNHHS EGVREKRDLI TKIISMREEE ECHRIVDPSS DKDVSQHKEK 660 DCLLQKLMKE KLYSWLLHKI MEDGKGPNVL DEKGQGVLHL AAALGYDWAI KPTVTAGVSI 720 NFRDANGWTA LHWAAFCGRE QTVAILVSLG AAAGAVTDPT PEFPLGRPPA DLASGNGHKG 780 ISGFLAESSL TSFLSNLTMN DQKEAVQTVS DRIATPVFDS DDILSLKDSL TAVCNATQAA 840 DRIHQMFRMQ SFQRKQLSES GDGVSDEHAI SLLTAKARRP LHIDGVAHAA ATQIQKKYRG 900 WKKRKEFLLI RQRIVKIQAH VRGHQVRKQY RTIVWSVGIL EKVILRWRRK GSGLRGFRRD 960 AITKEPDPQC TPSKEDDYDF LKEGRKQTEE RLQKALTRVK SMAQNPEGRG QYRRLLTLVQ 1020 GIRENKACDM VLSSTEEAGE GEGDGDEDFI DIETLLDDEN FMSIAFE* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, old leaves, petals, sepals, top of carpels, stigmas, stamen filaments, anthers and siliques, but not in pollen. {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
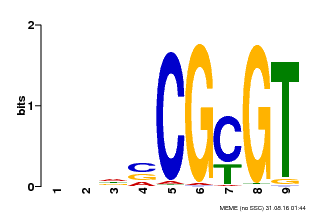
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012462697.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2-like isoform X3 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A0D2S9A0 | 0.0 | A0A0D2S9A0_GOSRA; Uncharacterized protein | ||||
| STRING | Gorai.013G061100.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Gorai.013G061100.1 |
| Entrez Gene | 105782485 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



