 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G003237.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 285aa MW: 32405.5 Da PI: 9.8106 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 98.9 | 2e-31 | 41 | 90 | 1 | 50 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeys 50
krien++nrqvtf+kRrng+lKKA+ELSvLCdaeva+i+fss+g+lyey+
HL.SW.v1.0.G003237.1 41 KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYA 90
79***********************************************8 PP
| |||||||
| 2 | K-box | 105.2 | 8.1e-35 | 108 | 205 | 3 | 100 |
K-box 3 kssgksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeen 91
+s++ s++e++a+ +qqe+akL+ +ie+L + +R llGe+++s + ++L++Le++Le+++++iRskKnell+++ie++qk+e +l+++n
HL.SW.v1.0.G003237.1 108 TSNTGSVSETNAQFYQQEAAKLRGQIESLHKGIRELLGECIDSAKPRDLKNLESKLERGISRIRSKKNELLFAEIEYMQKREIDLHNNN 196
4445559********************************************************************************** PP
K-box 92 kaLrkklee 100
+ Lr+k++e
HL.SW.v1.0.G003237.1 197 QLLRAKIAE 205
******986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 34.104 | 33 | 93 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 1.5E-42 | 33 | 92 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 5.21E-45 | 34 | 107 | No hit | No description |
| SuperFamily | SSF55455 | 2.75E-33 | 34 | 106 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 35 | 89 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 6.3E-33 | 35 | 55 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.2E-26 | 42 | 89 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 6.3E-33 | 55 | 70 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 6.3E-33 | 70 | 91 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 2.6E-25 | 116 | 203 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 14.371 | 119 | 209 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 285 aa Download sequence Send to blast |
MVNGSLNTNE VGKTMAYNQN NSMSMSVSPQ RKMGRGKIEI KRIENTTNRQ VTFCKRRNGL 60 LKKAYELSVL CDAEVALIVF SSRGRLYEYA NQSVKSTIDR YKKASSDTSN TGSVSETNAQ 120 FYQQEAAKLR GQIESLHKGI RELLGECIDS AKPRDLKNLE SKLERGISRI RSKKNELLFA 180 EIEYMQKREI DLHNNNQLLR AKIAENERNQ NMSMSMNINM SMNVGGGGGY ELMQQQQQQQ 240 SHEAQPAQPQ QQAFDSRDYF QVNALQPNNN HHYSRQDPMA LQLV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tqe_P | 5e-21 | 33 | 105 | 1 | 73 | Myocyte-specific enhancer factor 2B |
| 1tqe_Q | 5e-21 | 33 | 105 | 1 | 73 | Myocyte-specific enhancer factor 2B |
| 1tqe_R | 5e-21 | 33 | 105 | 1 | 73 | Myocyte-specific enhancer factor 2B |
| 1tqe_S | 5e-21 | 33 | 105 | 1 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_A | 5e-21 | 33 | 105 | 1 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_B | 5e-21 | 33 | 105 | 1 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_C | 5e-21 | 33 | 105 | 1 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_D | 5e-21 | 33 | 105 | 1 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_E | 5e-21 | 33 | 105 | 1 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_F | 5e-21 | 33 | 105 | 1 | 73 | Myocyte-specific enhancer factor 2B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the control of organ identity during the early development of flowers. Is required for normal development of stamens and carpels in the wild-type flower. Plays a role in maintaining the determinacy of the floral meristem. Acts as C class cadastral protein by repressing the A class floral homeotic genes like APETALA1. Forms a heterodimer via the K-box domain with either SEPALATTA1/AGL2, SEPALATTA2/AGL4, SEPALLATA3/AGL9 or AGL6 that could be involved in genes regulation during floral meristem development. Controls AHL21/GIK, a multifunctional chromatin modifier in reproductive organ patterning and differentiation (PubMed:19956801). Induces microsporogenesis through the activation of SPL/NZZ (PubMed:15254538). {ECO:0000269|PubMed:15254538, ECO:0000269|PubMed:19956801}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00609 | ChIP-seq | Transfer from AT4G18960 | Download |
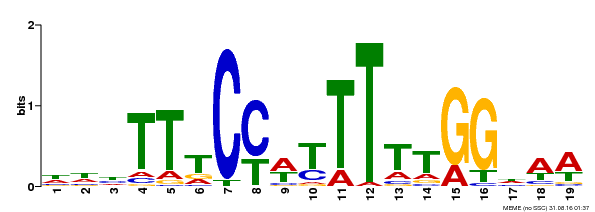
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by the A class floral homeotic protein APETALA2 and by other repressors like LEUNIG, SEUSS, SAP or CURLY LEAF. Positively regulated by both LEAFY and APETALA1. Repressed by silencing mediated by polycomb group (PcG) protein complex containing EMF1 and EMF2. Up-regulated by HUA2. {ECO:0000269|PubMed:10198637, ECO:0000269|PubMed:11058164, ECO:0000269|PubMed:1675158, ECO:0000269|PubMed:17794879, ECO:0000269|PubMed:18281509, ECO:0000269|PubMed:19783648, ECO:0000269|PubMed:9783581}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015876427.1 | 1e-134 | floral homeotic protein AGAMOUS isoform X3 | ||||
| Refseq | XP_015876435.1 | 1e-134 | floral homeotic protein AGAMOUS isoform X4 | ||||
| Swissprot | P17839 | 1e-120 | AG_ARATH; Floral homeotic protein AGAMOUS | ||||
| TrEMBL | A0A2P5C0X0 | 1e-144 | A0A2P5C0X0_PARAD; MADS-box transcription factor | ||||
| STRING | XP_008225406.1 | 1e-126 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF684 | 29 | 102 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18960.1 | 1e-113 | MIKC_MADS family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



