 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G005896.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1002aa MW: 111041 Da PI: 5.9304 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 131.5 | 2.9e-41 | 153 | 230 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C adl++ak+yhrrhkvCe+hska ++lv +++qrfCqqCsrfh+l+efDe+krsCrrrLa+hn+rrrk+++
HL.SW.v1.0.G005896.1 153 ICQVEDCGADLTSAKDYHRRHKVCEMHSKAVKALVGNVMQRFCQQCSRFHALQEFDEGKRSCRRRLAGHNKRRRKTNP 230
5**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 5.5E-33 | 147 | 215 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.975 | 151 | 228 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.09E-38 | 152 | 233 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.0E-29 | 154 | 227 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 6.53E-7 | 773 | 886 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.6E-6 | 774 | 887 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 3.09E-6 | 774 | 885 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1002 aa Download sequence Send to blast |
MEARFGGEPR HFFGMSSADL TTVRKRTMEW DLNQWKWDGD LFIASSVNPV VPSEGMGRQF 60 FPAGGSSNSN TSSSCSDEGN LVIEKGKREL EKRRRVNAVE DENLNDQTGN LTLKLGGRGH 120 HLHQPASERE MGNWEGTSGK KTKFVGGTSS RAICQVEDCG ADLTSAKDYH RRHKVCEMHS 180 KAVKALVGNV MQRFCQQCSR FHALQEFDEG KRSCRRRLAG HNKRRRKTNP DTVANGNSLS 240 DDQTSGYLLI TNKPDQSTEQ DLLSHLLKSL ANQTSEQGGK NVAGLLRDPQ NLLNDGASVG 300 NSEVVSTFLS NCSQGPPRPI KQNHAVSISE MPQQGIHLHD TNGGSVQATS SVKPSIMNSP 360 PSYSEARDST AGHSKMNNFD LNDIYIDSDD GVEDPERSFP PTNLVTSSLD CPSWVQQDSH 420 QSSPPQTNSD SASAQSPSSS NGEAQSRTDR IVFKLFGKEP NDFPLVLRAQ ILDWLSHSPS 480 DIESYIRPGC IILTIYLRQS ESAWEELCYD ISSKLSRLLD VSDDTFWSTG WTFIRVQHQI 540 AFVCNGQVIV DTSLPLRSSN CSKIVSIKPI ALPASEKAQF LVRGTNLVRP TMRLFCALEG 600 KYLVQEATHE LMDGDDNSEH DEQCINFSCP IEVTNGRGFI EIEDQGLGSS FFPFLVAEED 660 VCSEIRMLES VLEYTESDTV GDEIGKPESY NQAMDFIHEM GWLLHRSQLR SRLGDSDPNA 720 EPFTLKRFKW LMEFSVDHDW CAVVRKLLDI LIDGSVGEGE DRSLDLALSE IGLLHRAVRR 780 NSKALVEALL KYAPTNLSKL KSEEKSATDA PTPNFLFRPD VTGPGGLTPL HIAAGKDGSE 840 DVLDALTNDP GMVGIEAWKS ARDSTGSTPE DYARLRGHYT YIRLIQRKIN KKPGSGHVVV 900 DIPGDQKEDC SSNQKQNELA NTTTFEIGRS EQRRIQRPCG LCDRKLAYST SSRSVVYRPA 960 MLSMVAIAAV CVCVALLFKS SPEVLYVFRP FRWEMLEFGT S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-31 | 147 | 227 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 91 | 95 | KRRRV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
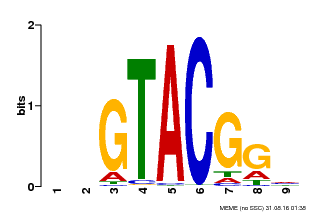
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010091413.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Refseq | XP_024018150.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | A0A2P5ETD2 | 0.0 | A0A2P5ETD2_TREOI; SBP-box transcription factor | ||||
| STRING | XP_010091413.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1539 | 34 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



