 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G007115.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 248aa MW: 27119.8 Da PI: 7.4579 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 75.6 | 8.3e-24 | 198 | 246 | 1 | 49 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSE CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrf 49
+Cq+egC+adls+ak+yhrrhkvCe+hska++v+++gl+qrfCqqCsr
HL.SW.v1.0.G007115.1 198 RCQAEGCNADLSHAKHYHRRHKVCEFHSKASTVIAAGLTQRFCQQCSRL 246
6**********************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 8.2E-25 | 193 | 245 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 20.529 | 196 | 247 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.09E-22 | 197 | 246 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.9E-17 | 199 | 246 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009554 | Biological Process | megasporogenesis | ||||
| GO:0009556 | Biological Process | microsporogenesis | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 248 aa Download sequence Send to blast |
MLDYEWGRVS AMMLTGEEHE QPPGSDPNRH NILDHYANAA AQTAFNDNLL AAQPGTNGSA 60 FANHHFNTTA THNQNHHHHH NHHHDSQAQV HSTHHVHPSL YDPRSYSGAS SYAPQHPSML 120 SLDPVLGGGG GGGGGHGGSG GFMLVPKSED MCRPPVDFSS RIGLNLGGRT YFSSEDDFMN 180 RLYRRPRPLD PASAHSPRCQ AEGCNADLSH AKHYHRRHKV CEFHSKASTV IAAGLTQRFC 240 QQCSRLF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj0_A | 2e-17 | 199 | 246 | 6 | 53 | squamosa promoter-binding protein-like 12 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. Binds specifically to the 5'-GTAC-3' core sequence. Involved in development and floral organogenesis. Required for ovule differentiation, pollen production, filament elongation, seed formation and siliques elongation. Also seems to play a role in the formation of trichomes on sepals. May positively modulate gibberellin (GA) signaling in flower. {ECO:0000269|PubMed:12671094, ECO:0000269|PubMed:16095614, ECO:0000269|PubMed:17093870}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00090 | SELEX | Transfer from AT1G02065 | Download |
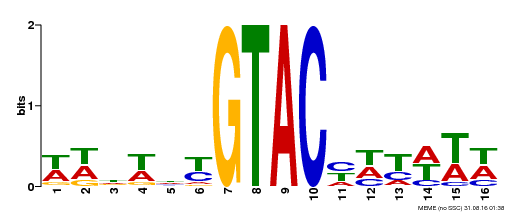
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007052207.2 | 8e-91 | PREDICTED: squamosa promoter-binding-like protein 8 | ||||
| Refseq | XP_015889429.1 | 7e-92 | squamosa promoter-binding-like protein 8 | ||||
| Refseq | XP_015889430.1 | 7e-92 | squamosa promoter-binding-like protein 8 | ||||
| Refseq | XP_024931908.1 | 7e-92 | squamosa promoter-binding-like protein 8 | ||||
| Swissprot | Q8GXL3 | 8e-51 | SPL8_ARATH; Squamosa promoter-binding-like protein 8 | ||||
| TrEMBL | A0A2P5C9P4 | 1e-120 | A0A2P5C9P4_TREOI; SBP-box transcription factor | ||||
| STRING | EOX96364 | 2e-90 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6801 | 33 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G02065.2 | 6e-49 | squamosa promoter binding protein-like 8 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



