 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G007775.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 393aa MW: 42949.2 Da PI: 6.8561 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 66.1 | 6.8e-21 | 186 | 235 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrd.psengkr.krfslgkfgtaeeAakaaiaarkkleg 55
+y+GVr+++ +g+W+AeIrd + + +r++lg+f+tae Aa+a+++a+++++g
HL.SW.v1.0.G007775.1 186 KYRGVRQRP-WGKWAAEIRDpF-----KaARVWLGTFDTAEAAARAYDEAALRFRG 235
8********.**********66.....56*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 8.2E-33 | 185 | 244 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.85E-23 | 186 | 244 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.3E-14 | 186 | 235 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.9E-36 | 186 | 249 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.38E-22 | 186 | 245 | No hit | No description |
| PROSITE profile | PS51032 | 25.686 | 186 | 243 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-11 | 187 | 198 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-11 | 209 | 225 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006970 | Biological Process | response to osmotic stress | ||||
| GO:0009749 | Biological Process | response to glucose | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 393 aa Download sequence Send to blast |
MCVLKVANSG DNKLIISNNN NNNGEYNWLN ESPLFYSGEP IMATGFGLMD REREMNAMVS 60 ALTRVVSGDS TDFTGRHHHH PNQWLEMSAS ASSDMGSGSG SGSYGNKRGR DEGGSDLSTE 120 SVSRLCRAFG DHPFPTQASN SALEETSYMW RSCTGPTQTT SFTSTYEYHQ EFPQRQQPEQ 180 EQPRRKYRGV RQRPWGKWAA EIRDPFKAAR VWLGTFDTAE AAARAYDEAA LRFRGNKAKL 240 NFPENVRLRS HPGNPAGTQL VVSDSTNTLL SVSTSAEPIV HSQVQLRSCT LSSSSDVVSR 300 NLQSYSPHVL NSERTMSLYD QMVLSQSVAS HVSSSSSLSP SASSSPFFSS SVSSSSPSLQ 360 LPSFSSPFWV EPPVQLRPAA SDSSPSNFPA EQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 6e-23 | 185 | 246 | 1 | 63 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00584 | DAP | Transfer from AT5G64750 | Download |
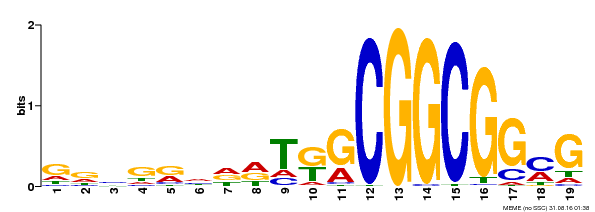
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM980447 | 2e-57 | KM980447.1 Betula platyphylla ethylene response factor protein 3 (ERF3) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024021449.1 | 1e-79 | ethylene-responsive transcription factor ABR1 | ||||
| TrEMBL | A0A2P5BG71 | 1e-140 | A0A2P5BG71_PARAD; AP2/ERF transcription factor | ||||
| STRING | XP_010096464.1 | 3e-69 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4933 | 32 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G43160.1 | 2e-20 | related to AP2 6 | ||||



