 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G008896.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 479aa MW: 51804.1 Da PI: 5.6533 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 126.2 | 9.8e-40 | 129 | 189 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++k+l+cprC+s++tkfCyynny+++qPr+fCkaC+ryWt+GG++rnvPvG+grrknk+s
HL.SW.v1.0.G008896.1 129 PDKILPCPRCNSMDTKFCYYNNYNVNQPRHFCKACQRYWTAGGTMRNVPVGAGRRKNKNSA 189
7899******************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 4.0E-31 | 122 | 186 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 2.6E-32 | 131 | 187 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.863 | 133 | 187 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 135 | 171 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 479 aa Download sequence Send to blast |
MLESKDPAIT LFGKKIPLPY EDEAPAVSGD DLASQPSERE VDNDEEEAEP EKDSSPRSVC 60 DEDSEQDGDL SPETDYSRKL DSLPESSSVN PKTPSINEES SHSKIEKTEK EQGDATNTNN 120 SQEKTLKKPD KILPCPRCNS MDTKFCYYNN YNVNQPRHFC KACQRYWTAG GTMRNVPVGA 180 GRRKNKNSAS HYRHISISEA LHAARIDASS NEAHHAALKT NGRVLSFGID APICDSMASV 240 LNLADKKVSS GNGNGFHKME GKENNGDDCS SVSSVTVSNS ADDIGRNCPQ EPQMHSINGQ 300 RFPSVPCLPG VPWPYPWNSG VPSPAFCPPG FPMSFYPAAY WNCAVPGAWN VPWFPPQPSS 360 PNQKSPSSGS GSNSPTLGKH SRDGDTLKPD SSENEEPPKQ KNGSVLVPKT LRIDDPSEAA 420 KSSIWATLGI KNDSISGGGM FKAFQSKSND MKGISEASPV LRANPAALSR SLNFHENS* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Regulates a photoperiodic flowering response. Transcriptional repressor of 'CONSTANS' expression. {ECO:0000250, ECO:0000269|PubMed:19619493}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00394 | DAP | Transfer from AT3G47500 | Download |
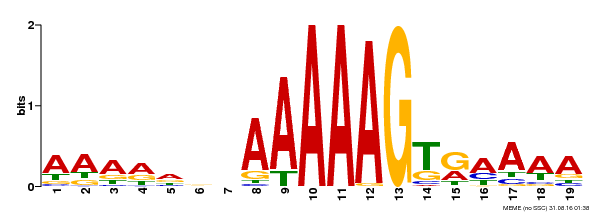
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Highly expressed at the beginning of the light period, then decreases, reaching a minimum between 16 and 29 hours after dawn before rising again at the end of the day. {ECO:0000269|PubMed:19619493}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010089911.1 | 0.0 | cyclic dof factor 3 | ||||
| Swissprot | Q8LFV3 | 1e-124 | CDF3_ARATH; Cyclic dof factor 3 | ||||
| TrEMBL | A0A2P5AJN0 | 0.0 | A0A2P5AJN0_PARAD; Zinc finger, Dof-type | ||||
| STRING | XP_010089911.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF625 | 34 | 147 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G47500.1 | 1e-111 | cycling DOF factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



