 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G009813.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 334aa MW: 36042.4 Da PI: 10.2557 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.1 | 9.8e-14 | 131 | 175 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ ++ + ++lG+g+W+ I+r + ++Rt+ q+ s+ qky
HL.SW.v1.0.G009813.1 131 PWTEEEHRTFLVGLEKLGKGDWRGISRSFVTTRTPTQVASHAQKY 175
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50158 | 8.532 | 3 | 18 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 17.905 | 124 | 180 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.87E-17 | 125 | 180 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 8.4E-18 | 127 | 179 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.9E-10 | 128 | 178 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.5E-11 | 128 | 174 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.3E-11 | 131 | 175 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.27E-9 | 131 | 176 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 334 aa Download sequence Send to blast |
MGRKCSHCGN IGHNSRTCTT SSSTATTFVN NNNNYANSSL RLFGVKVDIL SSSSSSAASS 60 SMNINALKKS FSMDCLMHSS LSSASSSPSS SLSSRIAPDE TISDHHKAAT NIGYLSDGLI 120 GRAQERKKGV PWTEEEHRTF LVGLEKLGKG DWRGISRSFV TTRTPTQVAS HAQKYFLRQA 180 TLNKKKRRSS LFDMVGISPL HSGSFRPSDD ESIRCSQHKD QNQQNGIVMP LLLELSSSKH 240 GQKLASDYNH QKPVVDFTQV ASHHPPNNSV PVWLHGLMDS QLKSKSNIIA SKPSSSNGAG 300 LDLELNLAGP RPSLEQNKPS NPGPLFVGPI TVT* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 183 | 187 | KKKRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00565 | DAP | Transfer from AT5G56840 | Download |
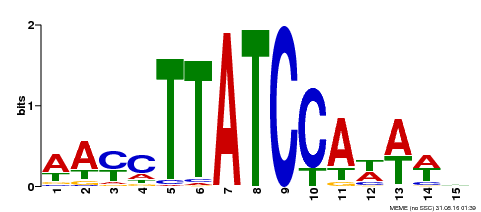
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023906593.1 | 1e-99 | transcription factor MYBS3-like isoform X2 | ||||
| TrEMBL | A0A2P5BNR2 | 1e-131 | A0A2P5BNR2_PARAD; GAMYB transcription factor | ||||
| STRING | EMJ03412 | 3e-94 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3213 | 32 | 71 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56840.1 | 2e-51 | MYB_related family protein | ||||



