 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G013013.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 312aa MW: 34488.1 Da PI: 8.3592 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.3 | 7.1e-18 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd++l+d+++++G g+W++ ++ g+ R++k+c++rw +yl
HL.SW.v1.0.G013013.1 14 KGAWTKEEDQRLIDYIRLHGEGCWRSLPKAAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 53.1 | 7.5e-17 | 67 | 111 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg++T+ Edel+++++ +lG++ W++Ia +++ gRt++++k++w+++
HL.SW.v1.0.G013013.1 67 RGNFTEIEDELIIKLHSLLGNK-WSLIAGRLP-GRTDNEIKNYWNTH 111
89********************.*********.************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 6.7E-24 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 17.586 | 9 | 61 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.9E-14 | 13 | 63 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 4.49E-29 | 13 | 108 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 4.4E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.49E-11 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 27.178 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.5E-27 | 65 | 117 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.1E-17 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.4E-15 | 67 | 111 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.01E-12 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0090378 | Biological Process | seed trichome elongation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 312 aa Download sequence Send to blast |
MGRSPCCEKA HTNKGAWTKE EDQRLIDYIR LHGEGCWRSL PKAAGLLRCG KSCRLRWINY 60 LRPDLKRGNF TEIEDELIIK LHSLLGNKWS LIAGRLPGRT DNEIKNYWNT HIKRKLISQG 120 LDPQTHRHLN ETAGDVAGAG PGAAALDFRN GSPAALPVAE INLMAEARQR KATVAPPNKS 180 VKESITETVL EDTNYCTSSG TTTEDDQRPE KEKPVFVQDQ RPVLNLELSI GIGPFQSRDT 240 AQSKLVHIIP QHQLFNGALQ SPNTAAVAAA VVSQSHGAVC LCCHLGYQAN ELCRNCQSTN 300 SFYRFHMPLD S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 1e-28 | 13 | 116 | 26 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00053 | PBM | Transfer from AT1G22640 | Download |
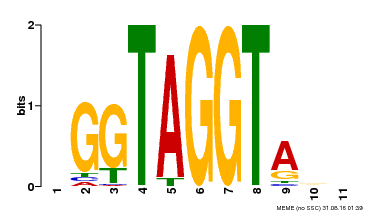
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By nitrogen, salicylic acid, NaCl and abscisic acid (ABA). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:18541146, ECO:0000269|PubMed:9839469}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010104477.1 | 1e-136 | transcription repressor MYB6 | ||||
| Swissprot | Q9S9K9 | 2e-94 | MYB3_ARATH; Transcription factor MYB3 | ||||
| TrEMBL | A0A2P5CWF2 | 1e-153 | A0A2P5CWF2_PARAD; MYB transcription factor | ||||
| TrEMBL | A0A2P5EHK4 | 1e-147 | A0A2P5EHK4_TREOI; MYB transcription factor | ||||
| STRING | XP_010104477.1 | 1e-136 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF264 | 34 | 221 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G22640.1 | 3e-96 | myb domain protein 3 | ||||



