 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G014778.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 669aa MW: 75057.9 Da PI: 6.3886 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 429.9 | 2.4e-131 | 33 | 356 | 1 | 303 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasd 89
eel++rmwkd+++lkrlker+k +++ a++++k +++++qarrkkmsraQDgiLkYMlk mevc+a+GfvYgiipekgkpv+gasd
HL.SW.v1.0.G014778.1 33 EELERRMWKDRIKLKRLKERQKLA--AQQ-AAEKQKPKQTSDQARRKKMSRAQDGILKYMLKLMEVCKARGFVYGIIPEKGKPVSGASD 118
8*******************9963..344.599******************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S-- CS
EIN3 90 sLraWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGke 178
++raWWkekv+fd+ngpaai+ky+a++l+l +++++ ++ ++++ l++lqD+tlgSLLs+lmqhc+ppqr+fplek v+pPWWPtG+e
HL.SW.v1.0.G014778.1 119 NIRAWWKEKVKFDKNGPAAIAKYEAECLALCEADNN--QNGNSQSILQDLQDATLGSLLSSLMQHCEPPQRKFPLEKKVPPPWWPTGNE 205
*******************************99999..5699999******************************************** PP
HHHHHHT--TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXXXX.. CS
EIN3 179 lwwgelglskdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsahss.. 265
+ww +lgl++ q ppykkphdlkk+wkv+vLtavikhmsp+i++ir+++rqsk+lqdkm+akes+++l vl++ee+++++ s+++
HL.SW.v1.0.G014778.1 206 DWWVKLGLPTGQT-PPYKKPHDLKKMWKVGVLTAVIKHMSPDIAKIRRHVRQSKCLQDKMTAKESAVWLGVLSREEALVRQPSSDNGts 293
*********9998.***********************************************************************4445 PP
.....XXXXXXXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXX....................X CS
EIN3 266 .....slrkqspkvtlsceqkedvegkkeskikhvqavktta....................g 303
r+++++ +++++++dv+g ++ + ++ + +++ +
HL.SW.v1.0.G014778.1 294 gvteiPPRRGEKDAAANSDSNYDVDGADDGLGSVSSRDPRRSqsmgversgnlqnntsqpgqD 356
788887788999*************88777755444433333445555555555544444440 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 8.0E-126 | 33 | 280 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.1E-70 | 154 | 288 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 2.09E-61 | 161 | 284 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 669 aa Download sequence Send to blast |
MAYEGDSSDI EVEDLRCDDL ADKDVSDEEI EPEELERRMW KDRIKLKRLK ERQKLAAQQA 60 AEKQKPKQTS DQARRKKMSR AQDGILKYML KLMEVCKARG FVYGIIPEKG KPVSGASDNI 120 RAWWKEKVKF DKNGPAAIAK YEAECLALCE ADNNQNGNSQ SILQDLQDAT LGSLLSSLMQ 180 HCEPPQRKFP LEKKVPPPWW PTGNEDWWVK LGLPTGQTPP YKKPHDLKKM WKVGVLTAVI 240 KHMSPDIAKI RRHVRQSKCL QDKMTAKESA VWLGVLSREE ALVRQPSSDN GTSGVTEIPP 300 RRGEKDAAAN SDSNYDVDGA DDGLGSVSSR DPRRSQSMGV ERSGNLQNNT SQPGQDKEQL 360 DKPPKRPRLN SKPTDQLPRP SQNEHLHIKP RNNLPDINHT EEPPKRPRLN SKPTDQLPQP 420 SQNEHLHIEP RNSLPDINHT EEVHLIGYHS EQNQQEDETV TGLRPPDKDM DVESQLPVSD 480 FNSLFALPSD NEVSTHSRHE GGAAMHFHEI PNTGIHHGDT YNFYNPSANY GPSHDRHHSP 540 VAINEIHVRP ADGVHLPTVH RNGSEISGGD LPYYVNNEQD RPVDSHFGSP LNSLSLDFGG 600 FNSPLNFGIN GTSVLDDLLL DDDLIQVVRY RLIVDWGSLH QSIIELISSP SICDYGSIRK 660 TNSIVGIM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 2e-72 | 160 | 291 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
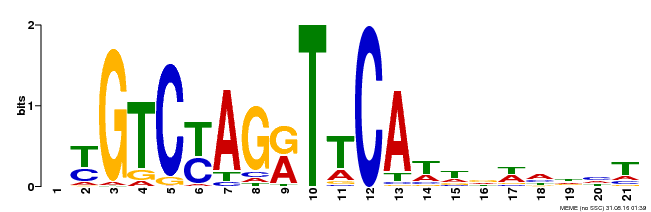
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010091992.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 0.0 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A2P5D8V8 | 0.0 | A0A2P5D8V8_PARAD; Ethylene insensitive 3-like protein, DNA-binding domain containing protein | ||||
| STRING | XP_010091992.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF322 | 34 | 200 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-172 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



