 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G017336.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 331aa MW: 36551.1 Da PI: 9.8857 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 42.5 | 1.5e-13 | 54 | 104 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqk 46
+++WT eE+e l +v+++G+g Wk I r +R+ ++k++w++
HL.SW.v1.0.G017336.1 54 KQKWTSEEEEALRAGVAKHGTGKWKDIQRDPEfnpflVNRSNIDLKDKWRN 104
79************************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.455 | 49 | 109 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.61E-17 | 51 | 107 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.4E-7 | 53 | 108 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.9E-9 | 54 | 104 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-15 | 54 | 105 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 1.84E-22 | 55 | 105 | No hit | No description |
| SMART | SM00526 | 1.5E-17 | 165 | 230 | IPR005818 | Linker histone H1/H5, domain H15 |
| PROSITE profile | PS51504 | 24.02 | 167 | 235 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene3D | G3DSA:1.10.10.10 | 2.8E-14 | 170 | 235 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.9E-15 | 170 | 240 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF00538 | 1.2E-10 | 171 | 228 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 331 aa Download sequence Send to blast |
MEAEPRLGLR LRIRVRVVAG RNLLKQEAAL VSPEAGTREK PRRESERRRM GNPKQKWTSE 60 EEEALRAGVA KHGTGKWKDI QRDPEFNPFL VNRSNIDLKD KWRNMSCSAN GQGPREKSRT 120 VKAKANSDAP ATQVSTPQTA VAVPAVRDSA TAPNADDSSK ALPDGKNSSM YNTMIFEALS 180 SLKEPNGSDT SAIVSYIEQR HEVPQNFRRL LSTRLRRLVA QDKLEKVQNC YKVKSHADAS 240 LGTKTPAPKP ESQPRQTQTT VYRTSTDVVD EAAVAAAYKV AEAENKSFVA AEAVKEADRI 300 SEMAEDAESL LQLAKEIFEK CSQGERVYAA * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
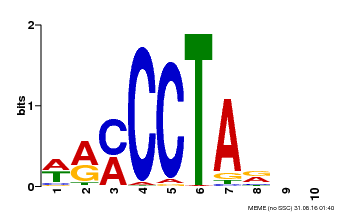
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010090083.2 | 1e-141 | telomere repeat-binding factor 4 | ||||
| Swissprot | F4IEY4 | 2e-74 | TRB5_ARATH; Telomere repeat-binding factor 5 | ||||
| TrEMBL | A0A2P5EFW3 | 1e-169 | A0A2P5EFW3_TREOI; Telomeric repeat-binding factor | ||||
| STRING | XP_010090083.1 | 1e-136 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3228 | 29 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G17520.1 | 7e-60 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



