 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G019443.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 583aa MW: 63796.3 Da PI: 7.4693 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48 | 2.8e-15 | 80 | 126 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT++Ede l av+ + g++Wk+Ia+ ++ R++ qc +rwqk+l
HL.SW.v1.0.G019443.1 80 KGGWTPQEDETLRSAVAAFNGKSWKKIAEFFN-DRSEVQCLHRWQKVL 126
688*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 60.5 | 3.7e-19 | 132 | 178 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd+++ ++v ++G W++Iar ++ gR +kqc++rw+++l
HL.SW.v1.0.G019443.1 132 KGPWTPEEDKKITELVSKYGATKWSLIARSLP-GRIGKQCRERWHNHL 178
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 52.5 | 1.2e-16 | 184 | 227 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+++WT +E++ l +a++ +G++ W+ Ia+ ++ gRt++ +k++w++
HL.SW.v1.0.G019443.1 184 KDAWTSDEELSLMHAHQIHGNK-WAEIAKVLP-GRTDNAIKNHWNS 227
679*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.947 | 75 | 126 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.5E-13 | 79 | 128 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.2E-14 | 80 | 126 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 2.23E-14 | 81 | 136 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.1E-17 | 82 | 134 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.57E-11 | 83 | 126 | No hit | No description |
| PROSITE profile | PS51294 | 31.279 | 127 | 182 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.61E-31 | 129 | 225 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.1E-17 | 131 | 180 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.8E-18 | 132 | 178 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.60E-16 | 134 | 178 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-27 | 135 | 185 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.949 | 183 | 233 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.2E-14 | 183 | 231 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.9E-15 | 184 | 227 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.57E-11 | 186 | 229 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 8.5E-23 | 186 | 233 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 583 aa Download sequence Send to blast |
MSFSSIPATL SPEEQKEMSE VKLEECCLEN KQSTAASSSS VSEGSGSVFL KSPGVCSPAA 60 STSPPQWYYP RTSGPVRRAK GGWTPQEDET LRSAVAAFNG KSWKKIAEFF NDRSEVQCLH 120 RWQKVLNPEL VKGPWTPEED KKITELVSKY GATKWSLIAR SLPGRIGKQC RERWHNHLNP 180 DIRKDAWTSD EELSLMHAHQ IHGNKWAEIA KVLPGRTDNA IKNHWNSSLK KKLDFFVSTG 240 SLPPATKNNS QNGPKDTNKC SATKKSLVCS NKEFDSTAHT LGNATMYTIN EDGNNQLDSS 300 GVLCSMLGSS SAPPNESADS EGIESKPVSS SMDPSYSNSD SVVKFENGGI NREANIENCL 360 ISKQSQHCGI NNEENGEVKE DKVVGTPLSS KTLFFGSLCY EPPQLDACIP LDSDFSSIHC 420 LQNEFNSSTY SSPGSCFTPP CVKGHGFTLP SPESVLKMAA LTFPNTPSIL RKRKYKAQTP 480 LLPNEIEKAD HATQEHDVSK TNLEMPGSQD GSLCKSPTYH GINSAGPNGK AFNASPPYRL 540 RSKRTAVLKS VEKQLEFTFD KEEHEGNVKS VEFSAKTKMG VT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 2e-65 | 79 | 233 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 2e-65 | 79 | 233 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (By similarity). Transcription repressor that regulates organ growth. Binds to the promoters of G2/M-specific genes and to E2F target genes to prevent their expression in post-mitotic cells and to restrict the time window of their expression in proliferating cells (PubMed:26069325). {ECO:0000250|UniProtKB:Q94FL9, ECO:0000269|PubMed:26069325}. | |||||
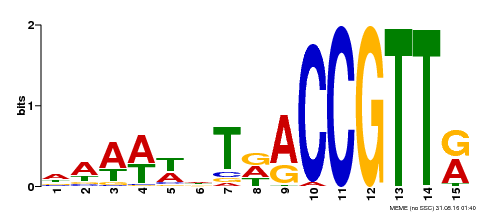
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by ethylene and salicylic acid (SA). {ECO:0000269|PubMed:16463103}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024031238.1 | 0.0 | transcription factor MYB3R-3 isoform X1 | ||||
| Swissprot | Q6R032 | 1e-154 | MB3R5_ARATH; Transcription factor MYB3R-5 | ||||
| TrEMBL | A0A2P5DS50 | 0.0 | A0A2P5DS50_TREOI; GAMYB transcription factor | ||||
| STRING | XP_010111464.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6616 | 34 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 1e-145 | myb domain protein 3r-5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



