 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G022516.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 282aa MW: 32556.8 Da PI: 5.3521 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.3 | 1.5e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT eEd++l+ +++++G g+W++ ++ g+ R++k+c++rw +yl
HL.SW.v1.0.G022516.1 14 KGPWTSEEDQILISYIQRYGHGNWRALPKQAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 54.5 | 2.7e-17 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T eE+e ++d++++lG++ W++Ia++++ gRt++++k+ w+++l
HL.SW.v1.0.G022516.1 67 RGNFTREEEETIIDLHEMLGNR-WSAIAARLP-GRTDNEIKNVWHTHL 112
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-26 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.992 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.83E-31 | 10 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.1E-14 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.5E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.34E-10 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 25.885 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.2E-27 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.3E-15 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-15 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.41E-10 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 282 aa Download sequence Send to blast |
MGRAPCCEKM GLKKGPWTSE EDQILISYIQ RYGHGNWRAL PKQAGLLRCG KSCRLRWINY 60 LRPDIKRGNF TREEEETIID LHEMLGNRWS AIAARLPGRT DNEIKNVWHT HLKKRLSTTT 120 TQNNKNHHVV QKSKRQRHQQ SCDDDHQVIK NDYDNDDDTK YNNNIMITMD QDEENNNNNN 180 RPVVSSPADQ SSSSSFSTGD HDHNDIINVM DDDFWSEVFS SGEIINDHNT SNSEVLLDGL 240 SVNNQFSDHA LITTTNLDYD QDMDFWYNIF TRSADLPELL L* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 5e-29 | 12 | 118 | 5 | 110 | B-MYB |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 131 | 136 | KSKRQR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00375 | DAP | Transfer from AT3G23250 | Download |
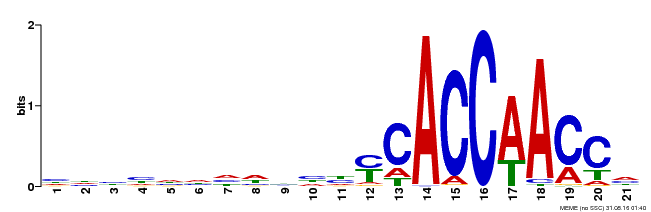
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015891267.1 | 4e-95 | transcription factor MYB13 | ||||
| TrEMBL | A0A2P5CNN2 | 1e-102 | A0A2P5CNN2_PARAD; MYB transcription factor | ||||
| STRING | cassava4.1_013493m | 2e-90 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G31180.1 | 1e-82 | myb domain protein 14 | ||||



