 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G025924.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 316aa MW: 34380.5 Da PI: 7.6941 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 31 | 5.8e-10 | 14 | 68 | 3 | 48 |
SS-HHHHHHHHHHHHHTTTT..........-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg..........tWktIartmgkgRtlkqcksrwqkyl 48
W++eE++ + +a++ + + +W + a++++ +++ +++k ++q++l
HL.SW.v1.0.G025924.1 14 TWSKEEEKAFENAIAIHWSTtdqedeeseeQWQKMASMVP-TKSMEELKRHYQMLL 68
7***************************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 38.8 | 2.1e-12 | 147 | 191 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ l++ + ++G+g+W++I+r + Rt+ q+ s+ qky
HL.SW.v1.0.G025924.1 147 PWTEEEHRLFLLGLDKFGKGDWRSISRNFVISRTPTQVASHAQKY 191
7*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 7.395 | 10 | 70 | IPR017884 | SANT domain |
| SMART | SM00717 | 1.8E-7 | 11 | 70 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.79E-7 | 13 | 71 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 3.1E-8 | 13 | 68 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.8E-4 | 14 | 69 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 7.01E-6 | 17 | 68 | No hit | No description |
| PROSITE profile | PS51294 | 16.873 | 140 | 196 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.61E-17 | 142 | 197 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.5E-17 | 144 | 194 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.3E-10 | 144 | 194 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.9E-11 | 145 | 190 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.14E-9 | 147 | 192 | No hit | No description |
| Pfam | PF00249 | 7.9E-11 | 147 | 191 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 316 aa Download sequence Send to blast |
MSSTSTTTTD ITTTWSKEEE KAFENAIAIH WSTTDQEDEE SEEQWQKMAS MVPTKSMEEL 60 KRHYQMLLDD VTAIEDGRIP VPYYTVPADS SKTMMGKDNS HVVSGSEKRM SSGHGSSGFS 120 GLQGHDSGGH GGKGVSRAEQ ERRKGIPWTE EEHRLFLLGL DKFGKGDWRS ISRNFVISRT 180 PTQVASHAQK YFIRLNSVNR DRRRSSIHDI TSVTNGEVPP HQAPITGVQQ PNTNSPSIPA 240 IGAPPVKHRG QTHMPGMGMY GAPMGHPVAA AAAAAAGHMA SAVGTPVMLP RGHHHHPPPY 300 VVPVAYPMAP PTMHQ* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to 5'-TATCCA-3' elements in gene promoters. Derepresses strongly the sugar-repressed transcription of promoters containing SRS or 5'-TATCCA-3' elements. Functions with GAMYB to integrate diverse nutrient starvation and gibberellin (GA) signaling pathways during germination of grains. Sugar, nitrogen and phosphate starvation signals converge and interconnect with GA to promote the co-nuclear import of MYBS1 and GAMYB, resulting in the expression of a large set of GA-inducible hydrolases, transporters, and regulators that are essential for mobilization of nutrient reserves in the endosperm to support seedling growth (PubMed:22773748). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:22773748}. | |||||
| UniProt | Transcription activator that binds to 5'-TATCCA-3' elements in gene promoters. Derepress strongly the sugar-repressed transcription of promoters containing SRS or 5'-TATCCA-3' elements. {ECO:0000250|UniProtKB:Q8LH59}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00191 | DAP | Transfer from AT1G49010 | Download |
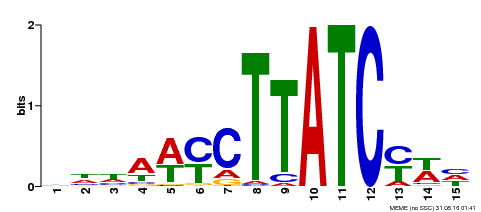
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by sucrose and gibberellic acid (GA). {ECO:0000269|PubMed:12172034}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007036438.2 | 1e-140 | PREDICTED: transcription factor DIVARICATA | ||||
| Swissprot | B8A9B2 | 3e-80 | MYBS1_ORYSI; Transcription factor MYBS1 | ||||
| Swissprot | Q8LH59 | 3e-80 | MYBS1_ORYSJ; Transcription factor MYBS1 | ||||
| TrEMBL | A0A2P5CM27 | 1e-155 | A0A2P5CM27_PARAD; GAMYB transcription factor | ||||
| STRING | EOY20939 | 1e-139 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF10694 | 33 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49010.1 | 2e-58 | MYB family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



