 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G026561.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 470aa MW: 51351 Da PI: 7.2067 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.2 | 1.6e-17 | 34 | 81 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+ Ed l ++vk++G g+W+++ ++ g++R++k+c++rw ++l
HL.SW.v1.0.G026561.1 34 KGPWTAAEDHVLMEYVKKHGEGNWNSVQKYSGLNRCGKSCRLRWANHL 81
79******************************************9986 PP
| |||||||
| 2 | Myb_DNA-binding | 51.8 | 1.8e-16 | 87 | 130 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++T+eE+ l++++++++G++ W++ a+ ++ gRt++++k++w++
HL.SW.v1.0.G026561.1 87 KGSFTPEEERLILELHAKYGNK-WARMAAQLP-GRTDNEIKNYWNT 130
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.059 | 29 | 81 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.2E-13 | 33 | 83 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 2.28E-31 | 33 | 128 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 6.6E-16 | 34 | 81 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.6E-24 | 35 | 88 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.40E-10 | 36 | 81 | No hit | No description |
| PROSITE profile | PS51294 | 26.49 | 82 | 136 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.8E-17 | 86 | 134 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.9E-16 | 87 | 130 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.1E-26 | 89 | 135 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.36E-13 | 89 | 130 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0090406 | Cellular Component | pollen tube | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 470 aa Download sequence Send to blast |
MTPHNNGGAQ SEGGSSGSSG GRCVSGPDGA ILKKGPWTAA EDHVLMEYVK KHGEGNWNSV 60 QKYSGLNRCG KSCRLRWANH LRPNLKKGSF TPEEERLILE LHAKYGNKWA RMAAQLPGRT 120 DNEIKNYWNT RVKRRKRQGL PLYPNDIQHL SPTSSLTPII QSPTTPTTPS FSFQSQNHVP 180 FSPTPPPHSP MSSPIQPFPN LTSPSSFEHN NSSFAFSSPS SFTFHRPVPV LGAPVRFKSF 240 RDSTGSGFSL PTQQSPSPPL GQYTPNQSTG SACFQFPLTF DPTLPQNIAT HFDSEHMGSS 300 GSIRSVKSEL PSSQFTQIHP EITCDNNNNS TNTNINNCNT KVISSATTQR NGGLLEDLLE 360 NLLEEAQRSV VSSAETVPKE ESKDQVDCLP DELSKLLNFV PGNLQAPEWY SDSSEISNGE 420 SLGVTDHNLC LDMQHLASLF PIASAADYGQ NPASWDNLPG ICKGWFKNN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 4e-32 | 32 | 135 | 5 | 107 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00287 | DAP | Transfer from AT2G32460 | Download |
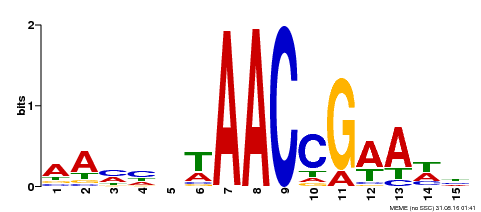
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010096901.1 | 1e-145 | transcription factor MYB97 | ||||
| TrEMBL | A0A2P5BRQ5 | 0.0 | A0A2P5BRQ5_PARAD; MYB transcription factor | ||||
| STRING | XP_010096901.1 | 1e-145 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G32460.1 | 2e-68 | myb domain protein 101 | ||||



