 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G030792.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1058aa MW: 119350 Da PI: 5.9635 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 179.8 | 3.2e-56 | 20 | 136 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahsee 90
++ ++rwl++ ei++iL+n++k++++ e+++ p++gsl+L++rk++ryfrkDG++w+kkkdgktv+E+he+LK g+++vl+cyYah+ee
HL.SW.v1.0.G030792.1 20 IEAQHRWLRPAEICEILQNYKKFRIAPEPANMPPNGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHERLKAGSIDVLHCYYAHGEE 108
5679************************************************************************************* PP
CG-1 91 nptfqrrcywlLeeelekivlvhylevk 118
n++fqrr+yw+Lee+l++ivlvhy+evk
HL.SW.v1.0.G030792.1 109 NENFQRRSYWMLEEDLSHIVLVHYREVK 136
*************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.92 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.4E-78 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.3E-49 | 21 | 134 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 4.1E-5 | 470 | 558 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 5.4E-6 | 473 | 557 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 2.13E-17 | 473 | 558 | IPR014756 | Immunoglobulin E-set |
| PROSITE profile | PS50297 | 17.263 | 638 | 777 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.0E-17 | 655 | 771 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 3.58E-16 | 663 | 768 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.29E-11 | 671 | 765 | No hit | No description |
| Pfam | PF12796 | 2.1E-6 | 678 | 769 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.041 | 706 | 735 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.084 | 706 | 738 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 840 | 745 | 774 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.12E-8 | 875 | 930 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.26 | 879 | 901 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.846 | 881 | 909 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0074 | 882 | 900 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 1.3E-5 | 902 | 924 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.889 | 903 | 927 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 5.4E-5 | 905 | 924 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1058 aa Download sequence Send to blast |
MADSRRLGLA NQLDIEQILI EAQHRWLRPA EICEILQNYK KFRIAPEPAN MPPNGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHER LKAGSIDVLH CYYAHGEENE NFQRRSYWML 120 EEDLSHIVLV HYREVKGNRT NYNRIKETEE ADTAPSSEID SPASSSIPPN SYQMLSQTTD 180 TTSLNSAQAS EYEDAESAYN QASSRLHPFL ELQQPFAEKI NAGINDAYYP VSFSNNNQEK 240 LSAIPGMDSS AGVSSDHPKN LDFPAWDNTL GNNAASIHLP FQPAFSATQS NNLTAIQKQE 300 QEPFEQLFTN GFGKHPQIQE EWQASEGNSR LPKWPVDQTL LTGSDNNLTF RYQSEVASGD 360 ELLKVHRGNA EHDDFVKPVS KSNMNLEEKP YISGMKQPLL DGSFAEEGLK KLDSFNRWMS 420 KELGDVNESH MQTTSRAYWN SVESDTCVDD SSQVRLDNYV LSPSLSQEQL FSIIDFSPNW 480 AYESSEVKVL ITGRFLKVDQ AEMNKWSCMF GEVEVPAEII AEGVLRCNAP IHKVGRVPFY 540 VTCSNRLACS EVREFEYRLN EARDVQTKYN QSGCTNEILK LRFGKLMSLT STSSNANPVN 600 LAEMSQLSNK ISSLLKEDQD EWDQMFRLTS EENFSVERVQ EQLHQKLLKE KLHEWLLHKV 660 AEGGKGPSLL DEGGQGVLHF AAALGYDWAL EPTIVAGVSV NFRDANGWTA LHWAAFCGRE 720 RTVASLISLG AAPGLLTDPS PKYPSGKTPA DLASDNGHKG ISGYLAESAL SAHLKCLDLD 780 AKAGKTTDTS ILNAVQTVSE RTATPIKDGD PDRLSLKDSL AAVCNATQAA ARIHQVFRVQ 840 SFQRKQLEEF SDDKFGLSNE QALSLIAVKA QKSGRQNDDV NAAAIRIQNK FRGWKGRKDF 900 LNMRQNVVKI QAHVRGHQVR KNYKKFVWSV GIVEKIILRW RRKGSGLRGF KSEPVVEVPS 960 IQDQSSSKDD DYDFLKEGRK QSEQRMQKAL ARVKSMVQYP EARNQYRRLL NVVTEIQEKK 1020 VPYDDVMYTE GTVDFEDDLI DIEALLEDDT YMPTATQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cxk_A | 3e-14 | 473 | 558 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_B | 3e-14 | 473 | 558 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_C | 3e-14 | 473 | 558 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_D | 3e-14 | 473 | 558 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_E | 3e-14 | 473 | 558 | 9 | 89 | calmodulin binding transcription activator 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
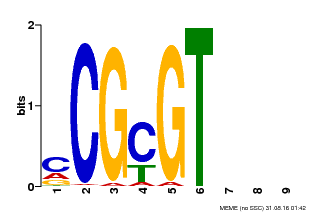
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024021686.1 | 0.0 | calmodulin-binding transcription activator 3 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A2P5BFW1 | 0.0 | A0A2P5BFW1_PARAD; Notch | ||||
| STRING | XP_010087599.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8284 | 27 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||



