 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G031956.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1051aa MW: 116325 Da PI: 8.5596 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.2 | 1.6e-40 | 134 | 211 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls ak+yhrrhkvCe hsk++++lv+ ++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+q+
HL.SW.v1.0.G031956.1 134 MCQVDNCKEDLSAAKDYHRRHKVCELHSKSTKALVASQMQRFCQQCSRFHPLSEFDEGKRSCRRRLAGHNRRRRKTQP 211
6**************************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.6E-33 | 129 | 196 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.778 | 132 | 209 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.62E-37 | 133 | 213 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.1E-29 | 135 | 208 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 6.13E-6 | 810 | 949 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 0.001 | 810 | 948 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.79E-4 | 853 | 952 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1051 aa Download sequence Send to blast |
MEEAGAQVAP PIFIHRTLSV MGKKRDLPYQ TPNFQQRFSN PNPVGNWNPK VWDWDTVRFL 60 AKPLDSGAAT INADHTKAEE PPVTPLTLTN KTPEVTDATL RLNLGGGFTS GDDPGSRPNK 120 RVRSGSPGST TYPMCQVDNC KEDLSAAKDY HRRHKVCELH SKSTKALVAS QMQRFCQQCS 180 RFHPLSEFDE GKRSCRRRLA GHNRRRRKTQ PDDVGSRLVL PGDRDNKING HLDIFNLLAA 240 VARAQGKNDD KNMNCSPILD KEQLVQILSK INSLPLPMDL AAKLHNLASL SRKIPDQTST 300 DHHDKLNGRA SQSTMDLLAA LSATLAPSAP DALATLSQRS SPSSDSGKTK LNCHDQANSS 360 VLQKRSPQEF PSLGGDRSST SYQSPMEDSD CQVQETRVNL PLQLFSSSPE NDSPPKLASS 420 RKYFSSDSSN PIEERSPSSS PPVMQTLFPM QTMAETVKSE KMSASRETNA NVESSRINGC 480 NMPFDLFRGS NRATEAGSIQ SVPHQAGYTS SGSDHSPSSL NSDVQDRTGR ILFKLFDKDP 540 SHFPGTLRTQ IFNWLSNSPS EMESYIRPGC VVLSVYVSMP SAAWEQLQEN LLQRLSSLVH 600 SSASDFWRNG RFLVHAGRQL ASHKDGKFRL CKSWKTWSSP ELISVTPLAV VGGQETSLIL 660 RGRNLNNLGT KIHCTDKGVY TTMEVRASTY QGTMYEEINL CGFRTHDASP SLFGRCFIEV 720 ENGFKGNSFP VIIADASICK ELRVLESVFY GEKKVSEVIA EDENHDDGRP RSKDEVLHFL 780 NELGWLFQRK KASSLLNGPH YSLGRFKFLL TFSVERNCSS LIKTLLDILV ERNMDENELS 840 REAVEMLSEV QLLHRAVKSK CKKMVDLLIN YSVIGSNDVS KKYIFQPNHV GPGYITPLHL 900 AACMSSSDEM IDALTNDPQE IGLNSWNSLL DANGQSPHAY ALMTNNQSYN KLVARKLGDR 960 TNGQITVTVG NAMSVEYQHG SRRSCAKCAV AATKYKRRVP GSQGLLQRPY VHSMLAIAAV 1020 CVCVCLFLRG LPDIGAVAPF KWENLDYGTI * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 7e-32 | 125 | 208 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
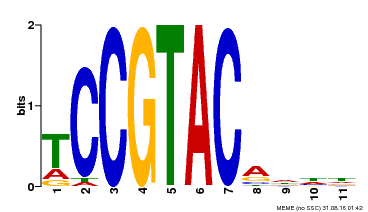
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010094493.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A2P5DIW9 | 0.0 | A0A2P5DIW9_TREOI; SBP-box transcription factor | ||||
| STRING | XP_010094493.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9694 | 31 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



