 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G032600.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 509aa MW: 55432.2 Da PI: 6.1403 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 124.9 | 2.6e-39 | 135 | 195 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++k+l+cprC+s++tkfCyynny+++qPr+fCk+C+ryWt+GG++rnvPvG+grrknk+s
HL.SW.v1.0.G032600.1 135 PDKILPCPRCNSMDTKFCYYNNYNVNQPRHFCKKCQRYWTAGGTMRNVPVGAGRRKNKNSA 195
7899******************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 4.0E-31 | 132 | 192 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 5.8E-32 | 137 | 193 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.733 | 139 | 193 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 141 | 177 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 509 aa Download sequence Send to blast |
MRAEMSEAKD PAIKLFGKTI PVAEVPGISG GSQGPSVSAC ATSVDDNTDQ EPASKPNSSS 60 EVNTSKDEED RETEKETAKG NPTETKEEDG NQPMSPEEPR NSDATSGISE NPKASKTEEE 120 QSEGTNSQEK TLKKPDKILP CPRCNSMDTK FCYYNNYNVN QPRHFCKKCQ RYWTAGGTMR 180 NVPVGAGRRK NKNSASHYRH ITVSEALQNA RTDVSTGVHS PHPAMKSNAT VLTFGSDAPL 240 CESMASVLNL ADKTMRNCTQ NGFHKPEELR TGFQKPEEVR TPVPYGSVES GDNNVDGSSV 300 TTSNSKDEAC KNVSQEQVIQ NSQGVPPQMP CFPGPPWPYP WNAAQWNPSV PMPAFCPPGY 360 PMPFYPAAAY WGCNVPGTWN IPWLPLPTSP NPVAPSSGPN SPTLGKHSRD ENTVKPNSSE 420 EEQPKESSSE RYLWIPKTLR IDDPGEAARS SIWATLGIKN DKADSISGGG LFKAFQPKND 480 EKNRRAEHSV LQANPAALSR SLSFQERS* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00046 | PBM | Transfer from AT5G39660 | Download |
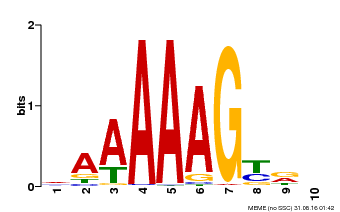
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010088142.1 | 0.0 | cyclic dof factor 2 | ||||
| TrEMBL | A0A2P5FF12 | 0.0 | A0A2P5FF12_TREOI; Zinc finger, Dof-type | ||||
| STRING | XP_010088142.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF625 | 34 | 147 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G39660.2 | 1e-107 | cycling DOF factor 2 | ||||



