 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G038994.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 953aa MW: 105013 Da PI: 5.0004 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 78.8 | 5.5e-25 | 147 | 248 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvF 82
f+k+lt sd++++g +++p++ ae+ + ++++ ++l+++d++ ++W++++iyr++++r++lt+GW+ Fv a++L++gD+v+F
HL.SW.v1.0.G038994.1 147 FCKTLTASDTSTHGGFSVPRRAAEKLfppldytM-QPPN-QELVVRDLHENTWTFRHIYRGQPKRHLLTTGWSLFVGAKRLRAGDSVLF 233
99*********************999*****954.5444.49*********************************************** PP
EE-SSSEE..EEEEE-S CS
B3 83 kldgrsefelvvkvfrk 99
++++ +l+v+++r+
HL.SW.v1.0.G038994.1 234 I--RDEKSQLMVGIRRA 248
*..4577778*****97 PP
| |||||||
| 2 | Auxin_resp | 113 | 2.8e-37 | 273 | 356 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+++s+F+++YnPra+++eFv+++ +++ka++ +++svGmRf m+fete+s +rr++Gt+vg+s+ldp+rWp+SkWr+L+
HL.SW.v1.0.G038994.1 273 AAHAAANRSPFTIFYNPRACPAEFVIPLTRFRKAVYaTQLSVGMRFGMMFETEESGKRRYMGTIVGISELDPLRWPGSKWRNLQ 356
79*********************************989********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 7.85E-47 | 134 | 276 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 4.1E-42 | 140 | 261 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.34E-23 | 146 | 247 | No hit | No description |
| PROSITE profile | PS50863 | 13.169 | 147 | 249 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.4E-26 | 147 | 249 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.0E-22 | 147 | 248 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 3.1E-32 | 273 | 356 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 3.4E-10 | 836 | 925 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 25.619 | 838 | 922 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 2.55E-9 | 851 | 917 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0010305 | Biological Process | leaf vascular tissue pattern formation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048507 | Biological Process | meristem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 953 aa Download sequence Send to blast |
MSSVEEKIKN GVLVAGASTN LLEEMKLLKE MQDQNGSRKS ISSELWHACA GPLVSLPQVG 60 SLVYYFPQGH SEQVAVSTKR LATSQIPNYP NLPSQLLCQV QNVTLHADRE TDEIYAQMSL 120 QPVNSEKDVF PVPDFGLKPS KHPSEFFCKT LTASDTSTHG GFSVPRRAAE KLFPPLDYTM 180 QPPNQELVVR DLHENTWTFR HIYRGQPKRH LLTTGWSLFV GAKRLRAGDS VLFIRDEKSQ 240 LMVGIRRANR QQSTLPSSVL SADSMHIGVL AAAAHAAANR SPFTIFYNPR ACPAEFVIPL 300 TRFRKAVYAT QLSVGMRFGM MFETEESGKR RYMGTIVGIS ELDPLRWPGS KWRNLQVEWD 360 EPGCCDKQNR VSSWDIETPE SLFIFPSLTS GLKRPFHSGF LDTDWGNMIK RPFNRGPENG 420 NGDLLPYGIS NLYSEQLIKM LLKPQLVNYP GTLSSLQQES AMAIADPMRD AKLMQTTLNQ 480 KDQVICSEGS SLQQNEMNPP QSTLDQSGVS NMNSSPNGNL HKNFNTAAKL DNQKVGGVIN 540 TEKTKVETEL STDQLSQLTS STVHSSEEKL ATGLVSSQSL VSQLTHFNQN QGPVQLQANQ 600 WGMQPQLDSL IYLSQQTDLN PSDMNNTNVS LPSLDPDECM YYSSCHPYSA GILRSPGSLP 660 GFGFQDSSAF SEAHNLPLPS MGQDVWDNNN LKVQAEHLTS FAHQEPCNFS SLTNSSNIKD 720 LSDESNNQSG IYGCPNIDGS NGASLAVDPS ASSTILDEFS TLKNVDFQNP SDCLVGNFST 780 SQDVQSQITT ASLGDSQAFS RQDIQDNSGG TSSSNVDLDE CSLLQNNNSW QQVVPPVRTY 840 TKVQKTGSVG RSIDVSSFKN YDELCSAIEC MFGLEGLLND PRGSRWKLVY VDYENDVLLV 900 GDDPWEEFVG CVRCIRILSP SEVQQMSEEG MKLLNSAAMQ GINAPVTEVG RA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 0.0 | 1 | 381 | 5 | 392 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Mediates embryo axis formation and vascular tissues differentiation. Functionally redundant with ARF7. May be necessary to counteract AMP1 activity. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:14973283, ECO:0000269|PubMed:17553903}. | |||||
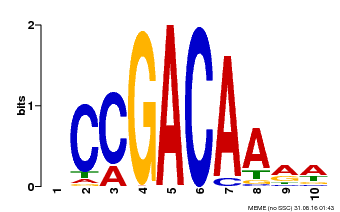
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00153 | DAP | Transfer from AT1G19850 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010095167.1 | 0.0 | auxin response factor 5 | ||||
| Refseq | XP_024020560.1 | 0.0 | auxin response factor 5 | ||||
| Swissprot | P93024 | 0.0 | ARFE_ARATH; Auxin response factor 5 | ||||
| TrEMBL | A0A2P5F7V6 | 0.0 | A0A2P5F7V6_TREOI; Auxin response factor | ||||
| STRING | XP_010095167.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8991 | 32 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19850.1 | 0.0 | ARF family protein | ||||



