 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G041795.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 850aa MW: 94547.4 Da PI: 6.9366 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 72.6 | 4.9e-23 | 174 | 275 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFk 83
f+k+lt sd++++g +++ +++a+e+ + + + +++l+ +d+++++W++++i+r++++r++l++GW+ Fv++++L +gD ++F
HL.SW.v1.0.G041795.1 174 FCKTLTASDTSTHGGFSVLRRHADEClppldmSRQ-PPTQELVAKDLHSNEWRFRHIFRGQPRRHLLQSGWSVFVSSKRLVAGDAFIFL 261
99*****************************7333.34459************************************************ PP
E-SSSEE..EEEEE-S CS
B3 84 ldgrsefelvvkvfrk 99
+ +++el+v+v+r+
HL.SW.v1.0.G041795.1 262 --RGENGELRVGVRRA 275
..449999*****996 PP
| |||||||
| 2 | Auxin_resp | 119.9 | 2e-39 | 300 | 382 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha+st+++F+v+Y+Pr+s++eF+v+++++++a+k+++s+GmRfkm+fe+e+++e+r++Gt++g++++dp rW++SkWr+Lk
HL.SW.v1.0.G041795.1 300 AWHAISTGTMFTVYYKPRTSPAEFIVPCDQYLEATKNNYSIGMRFKMKFEGEEAPEQRFTGTIIGIEEFDPQRWKDSKWRCLK 382
89*******************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 8.8E-41 | 170 | 289 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.03E-39 | 170 | 303 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.07E-20 | 172 | 274 | No hit | No description |
| PROSITE profile | PS50863 | 11.491 | 174 | 276 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 8.0E-21 | 174 | 275 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 7.9E-23 | 174 | 276 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 1.7E-36 | 300 | 382 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 1.1E-12 | 707 | 813 | IPR033389 | AUX/IAA domain |
| SuperFamily | SSF54277 | 1.08E-11 | 722 | 801 | No hit | No description |
| PROSITE profile | PS51745 | 25.592 | 724 | 808 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008285 | Biological Process | negative regulation of cell proliferation | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010227 | Biological Process | floral organ abscission | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 850 aa Download sequence Send to blast |
MASSEASVKG NSGNPMGESF TSGYTDHNDA RKAAEARKGF QFLTRFSEDF SVLMSFLLLR 60 VVDAEAALYE ELWHACAGPL VTVPRKGELV FYFPQGHIEQ VEASTNQVAD QQMPVYNLPP 120 KILCRVMNVE LKAEPDTDEV FAQITLMPVA NQNDNALEKE TPPPPPPRFQ VHSFCKTLTA 180 SDTSTHGGFS VLRRHADECL PPLDMSRQPP TQELVAKDLH SNEWRFRHIF RGQPRRHLLQ 240 SGWSVFVSSK RLVAGDAFIF LRGENGELRV GVRRAMRKQE NVPSSVISSH SMHLGVLATA 300 WHAISTGTMF TVYYKPRTSP AEFIVPCDQY LEATKNNYSI GMRFKMKFEG EEAPEQRFTG 360 TIIGIEEFDP QRWKDSKWRC LKVRWDETSA IPRPERVSPW KIEPALAPPA LNPLPVPRSK 420 RPRSNIVPLS PDSSVLTRDG SSKVTVDPSL PSGFSRVLQG QELSTLRGNF AESNDSDTAE 480 KSVVWQPSLD DEKIDVVSAS RRYVPENWAS SGKHEPTYTD LLSGFGATVD STRGFCTSFT 540 DHSVATANSM RKHSLDKEGR LNLHSSPRSM VPSSLSLSLD TNMKGSVQGG ISFQGQGNVR 600 YGGFDDYTAL HGHRVEHSHG NWFMPPPSSP HYENSVHSRE LTSKPVSEQK YEAVKPKDGN 660 CKIFGYSLIA PEPAVSHKTI VGKARGTKIA ADSPMASSEQ EKPLQTNQQH SREHQGKAQN 720 GSTRSCTKVH KQGIALGRSV DLTKFNNYEE LIAELDQLFE FGGELMAPKK NWLIVYTDDE 780 GDMMLVGDDP WQEFCGMVRK IFIYTREEVQ KMSPGTLNAH GDGNPSVCEV MDAKEKSQTL 840 PLSSVPENC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-177 | 67 | 404 | 18 | 354 | Auxin response factor 1 |
| 4ldw_A | 1e-177 | 67 | 404 | 18 | 354 | Auxin response factor 1 |
| 4ldw_B | 1e-177 | 67 | 404 | 18 | 354 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Could act as transcriptional activator or repressor. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Promotes flowering, stamen development, floral organ abscission and fruit dehiscence. Functions independently of ethylene and cytokinin response pathways. May act as a repressor of cell division and organ growth. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:15960614, ECO:0000269|PubMed:16176952, ECO:0000269|PubMed:16339187}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00574 | DAP | Transfer from AT5G62000 | Download |
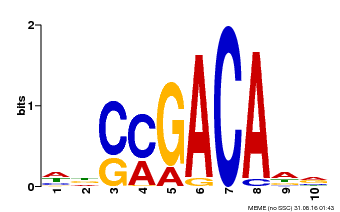
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024023134.1 | 0.0 | auxin response factor 2B isoform X1 | ||||
| Refseq | XP_024023135.1 | 0.0 | auxin response factor 2B isoform X2 | ||||
| Refseq | XP_024023136.1 | 0.0 | auxin response factor 2B isoform X3 | ||||
| Swissprot | Q94JM3 | 0.0 | ARFB_ARATH; Auxin response factor 2 | ||||
| TrEMBL | A0A2P5FI45 | 0.0 | A0A2P5FI45_TREOI; Auxin response factor | ||||
| STRING | XP_010099050.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3281 | 33 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62000.3 | 0.0 | auxin response factor 2 | ||||



