 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | HL.SW.v1.0.G044686.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Cannabaceae; Humulus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 287aa MW: 32635.1 Da PI: 5.7858 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 64.8 | 1.8e-20 | 135 | 185 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+y+GVr+++ +g+++AeIrd s ng +r++lg+f+taeeAa a+++a+++++g
HL.SW.v1.0.G044686.1 135 HYRGVRRRP-WGKYAAEIRDSSRNG--ARVWLGTFDTAEEAALAYDKAALRIRG 185
7********.**********55465..*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 2.5E-31 | 134 | 194 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.48E-30 | 135 | 193 | No hit | No description |
| Pfam | PF00847 | 1.3E-14 | 135 | 185 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.11E-23 | 135 | 195 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 23.894 | 135 | 193 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 8.9E-38 | 135 | 199 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.7E-11 | 136 | 147 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.7E-11 | 159 | 175 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 287 aa Download sequence Send to blast |
MVAIHKGQQS EAILENVWAS FISEKEEVIN EGKRLSKSWD DLPHLDGKEG SMEVLQRLPS 60 LGRWISMGAE TWEELLDSII VDPSRNLDRH DQHQQHSSTN NNTTANDDID HDKESNSVER 120 AKKKTAAMVV AATRHYRGVR RRPWGKYAAE IRDSSRNGAR VWLGTFDTAE EAALAYDKAA 180 LRIRGTKAYL NFPLETVAKS NNKTTTATAT TRCGKRESRE WENHQINYDH DQVMMTMMIN 240 QEPALRRSKA SSHDHENNNN DDPFDVFEFQ DLGSDYLDSL LSFPCE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 8e-30 | 131 | 196 | 1 | 66 | ATERF1 |
| 3gcc_A | 8e-30 | 131 | 196 | 1 | 66 | ATERF1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00441 | DAP | Transfer from AT4G18450 | Download |
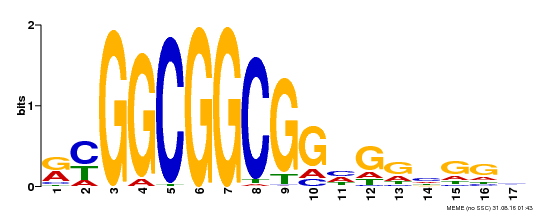
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024031507.1 | 4e-92 | ethylene-responsive transcription factor 1B | ||||
| TrEMBL | A0A2P5EBP2 | 1e-119 | A0A2P5EBP2_TREOI; AP2/ERF transcription factor | ||||
| STRING | EOY28113 | 2e-86 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9547 | 31 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18450.1 | 2e-27 | ERF family protein | ||||



