 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | IGS.gm_8_00085 | ||||||||
| Common Name | CHLNCDRAFT_144757 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Chlorellales; Chlorellaceae; Chlorella
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 473aa MW: 49932.7 Da PI: 8.5436 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 33.3 | 8.8e-11 | 139 | 185 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h ++E rRR+riN+++e Lr l+P+ ++ + a Le++v+Y++ Lq
IGS.gm_8_00085 139 MHVQAEARRRSRINERLEALRLLVPHT-----ERANTANFLEEVVQYVQRLQ 185
7999**********************8.....7******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 12.608 | 135 | 184 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.83E-11 | 138 | 197 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 6.0E-8 | 139 | 185 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.9E-12 | 139 | 195 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.70E-7 | 140 | 188 | No hit | No description |
| SMART | SM00353 | 1.3E-6 | 141 | 190 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 473 aa Download sequence Send to blast |
MTKEPRLCTG AASTSPLALD PNALTALQDL LARVPAAAVN MQAASAMPFV SHQQDLEVQQ 60 LLQHLQSSAR RERDAGAPQL PVPPALPNGG DGGAPPAPSR PAAATSRPGR SAGEARPATS 120 YNARHQQAPS SRSVLFHAMH VQAEARRRSR INERLEALRL LVPHTERANT ANFLEEVVQY 180 VQRLQSRVTD LERQLGLPAS VAAGQKAIAF ADDTPEATTL TAQQDDRRAS QSMPEQGPAP 240 PAQPTQALPA STPPAPAPEA QAGVHSVQAV LALLQQASQA PPVPAQEQVQ MLLLQAMQAQ 300 AVQAQVQQQL DLLQQQQQQQ VLPRHQPAAQ FSLQFQPPPS LPQQQQQQQQ QQQPQAQAFL 360 QGMGSRPLAH SGATSGGGSG SRGDSGARRP YPAALGPQML AAAESLGTLS AGGGSRRSAS 420 RSVVQEEADA AAAEAAKRRG GPGIATDEEL DQLHVHLAGQ QDDAKRRRLE SQ* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 464 | 468 | KRRRL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00219 | DAP | Transfer from AT1G69010 | Download |
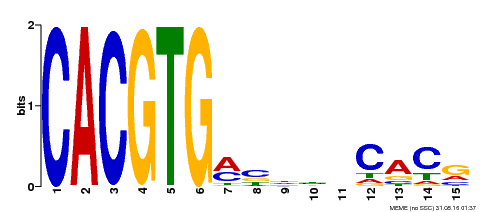
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_005848230.1 | 0.0 | hypothetical protein CHLNCDRAFT_144757 | ||||
| TrEMBL | E1ZCY3 | 0.0 | E1ZCY3_CHLVA; Uncharacterized protein | ||||
| STRING | XP_005848230.1 | 0.0 | (Chlorella variabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP5665 | 8 | 8 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 4e-10 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 17355663 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



