 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc000002.1_g00064.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 498aa MW: 53532.2 Da PI: 4.8713 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 124.9 | 2.7e-39 | 137 | 197 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++k+l+cprC+s++tkfCy+nny+++qPr+fCk+C+ryWt+GG++rnvPvG+grrknk+s
Itr_sc000002.1_g00064.1 137 PDKILPCPRCNSMETKFCYFNNYNVNQPRHFCKNCQRYWTAGGTMRNVPVGAGRRKNKNSG 197
7899******************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 2.0E-30 | 130 | 194 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 1.2E-31 | 139 | 195 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.789 | 141 | 195 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 143 | 179 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 498 aa Download sequence Send to blast |
MSDSSKDPAI KLFGKTIQLP EIPASTAPAT EGSCPDDAPG DDSSARHGSG SGNDPAEDGG 60 GGEEDEHLQK NQNGGKLDES KEDESLMAEE LTDQTNEMET DELKMPSNNK DCGAMKKSEG 120 EEQGETGNSQ EKALKKPDKI LPCPRCNSME TKFCYFNNYN VNQPRHFCKN CQRYWTAGGT 180 MRNVPVGAGR RKNKNSGSHY RHVTISEPFP SARVDLPNGI QLPTINPNGT LLTFGSDTPL 240 CESMASVLNI AEKTMQNGSI NGFHKLEELG VPVPHGAGDN GDDRSSGSSV TAASTKDESV 300 KNEVPGRQNC PNFAPQLPCF PGAPWPYPLT AVQWSSAIPP PGYCPPGFPM PVYPAPAYWG 360 CAVPGSWSVP WVSPPIAPQH HMPSTSGPNS PTLGKHSRDE NMLKPVNGEG EDPKKESNPE 420 KCLWVPKTLR IDDPEDAAKS SIWATLGIKN DKADSVGSGL FKAFQSKDDE KTDASENSTV 480 LQANPAALSR SLNFHENS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00046 | PBM | Transfer from AT5G39660 | Download |
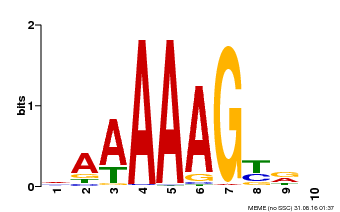
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019185496.1 | 0.0 | PREDICTED: cyclic dof factor 2 | ||||
| Refseq | XP_019185497.1 | 0.0 | PREDICTED: cyclic dof factor 2 | ||||
| TrEMBL | A0A068U1T1 | 0.0 | A0A068U1T1_COFCA; Uncharacterized protein | ||||
| STRING | VIT_14s0108g00980.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA532 | 24 | 122 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G39660.2 | 1e-113 | cycling DOF factor 2 | ||||



