 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc000038.1_g00006.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 391aa MW: 42640.5 Da PI: 6.5115 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 31.5 | 3.3e-10 | 238 | 286 | 3 | 55 |
HHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 3 rahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR+ri +++ L+el+P + +k Ka +L + ++Y++sLq
Itr_sc000038.1_g00006.1 238 NSHSLAERVRRERISERMRLLQELVPGC----NKITGKAVMLDEIINYVQSLQ 286
58**************************....999*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 5.23E-17 | 233 | 303 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.50E-11 | 233 | 290 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 2.5E-16 | 235 | 303 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 15.472 | 235 | 285 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.8E-7 | 238 | 286 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.1E-10 | 241 | 291 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 391 aa Download sequence Send to blast |
MGTEDNNNGD MGGLQHRGNV NISMAMSSES TFRSPTGMDS LPFCGSGWDP MVPVNQSLHY 60 PVAIDNQPMS STSHLVHHYP SGSDVVDMVP KVPAFGNEGF SDMVSSFGLP DCRLNYMHNI 120 GVGTNRAFSR GQHSQEDCLN LEEAAIGASP LGKKRRSSEA LSPSIASKNV EREQPKDPLG 180 DNSECSKEDE KKQKTEQNNS SSLRSKQAGK QVKDTSSSGE PPKENYVHVR AKRGQATNSH 240 SLAERVRRER ISERMRLLQE LVPGCNKITG KAVMLDEIIN YVQSLQQQVE FLSMKLATVN 300 PELNFDIERI LSKDILHAQG SNGALLGLGP GLNSPHPFPG APLGTFSNVP NTTPFHSLPQ 360 NVWENDLQSI LQMGFDSNSS LNNMGPNGNR S |
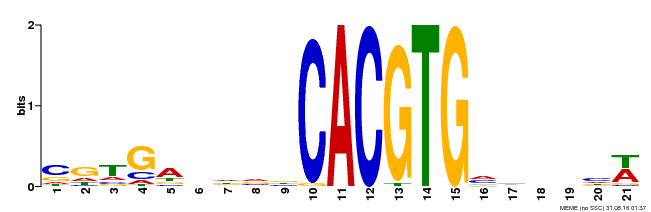
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00135 | DAP | Transfer from AT1G10120 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019174884.1 | 0.0 | PREDICTED: transcription factor bHLH74-like isoform X1 | ||||
| TrEMBL | A0A314L7E0 | 1e-156 | A0A314L7E0_NICAT; Transcription factor bhlh74 | ||||
| STRING | XP_009590631.1 | 1e-153 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA5250 | 24 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G10120.1 | 7e-67 | bHLH family protein | ||||



