 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc000078.1_g00028.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 263aa MW: 29768 Da PI: 9.5007 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 98.7 | 2.4e-31 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krienk+nrqvtf+kRrng+lKKA+ELS+LCdaeva+iifs++gklye++s
Itr_sc000078.1_g00028.1 9 KRIENKINRQVTFAKRRNGLLKKAYELSILCDAEVALIIFSNRGKLYEFCS 59
79***********************************************96 PP
| |||||||
| 2 | K-box | 80.8 | 3.2e-27 | 81 | 159 | 8 | 86 |
K-box 8 sleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkeke 86
++++++++s +qe+ kLk+ +e Lq++qRhllGedL +L +k+L+qLe+q+++slk+iRs++++++l+q++el +k +
Itr_sc000078.1_g00028.1 81 NQTSKDSQSNYQEYLKLKAGVEVLQQSQRHLLGEDLVQLGIKDLEQLENQVDTSLKSIRSTRTKHMLDQLSELHQKVGM 159
3577889*******************************************************************99765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 33.543 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 5.0E-42 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.72E-45 | 2 | 78 | No hit | No description |
| SuperFamily | SSF55455 | 3.01E-34 | 2 | 85 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.9E-33 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 2.7E-26 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.9E-33 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.9E-33 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 5.5E-22 | 83 | 160 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 13.849 | 87 | 200 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010076 | Biological Process | maintenance of floral meristem identity | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048441 | Biological Process | petal development | ||||
| GO:0048442 | Biological Process | sepal development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 263 aa Download sequence Send to blast |
MGRGKVELKR IENKINRQVT FAKRRNGLLK KAYELSILCD AEVALIIFSN RGKLYEFCSS 60 SSMSKTLERY HRCSYGALEA NQTSKDSQSN YQEYLKLKAG VEVLQQSQRH LLGEDLVQLG 120 IKDLEQLENQ VDTSLKSIRS TRTKHMLDQL SELHQKVGML GFLRLGIQHF ENNIMNIKVE 180 QNLLEVNKAL RIKLGESNAA FLTPWEPQGQ NMQYGQHSSS QPIQPQQCHN TVQIGYNPTS 240 MQANNNAGTS TMNGPGVLPG WML |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 3e-20 | 1 | 70 | 1 | 69 | MEF2C |
| 5f28_B | 3e-20 | 1 | 70 | 1 | 69 | MEF2C |
| 5f28_C | 3e-20 | 1 | 70 | 1 | 69 | MEF2C |
| 5f28_D | 3e-20 | 1 | 70 | 1 | 69 | MEF2C |
| 6byy_A | 4e-20 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| 6byy_B | 4e-20 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| 6byy_C | 4e-20 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| 6byy_D | 4e-20 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| 6bz1_A | 4e-20 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| 6bz1_B | 4e-20 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| 6bz1_C | 4e-20 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| 6bz1_D | 4e-20 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable MADS-box transcription factor that functions with J2 and EJ2 in meristem maturation. {ECO:0000269|PubMed:28528644}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00030 | SELEX | Transfer from AT2G03710 | Download |
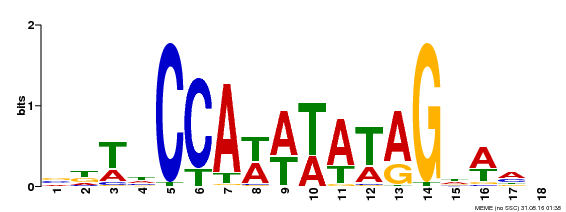
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019180616.1 | 1e-159 | PREDICTED: MADS-box protein CMB1-like | ||||
| Refseq | XP_019180617.1 | 1e-159 | PREDICTED: MADS-box protein CMB1-like | ||||
| Refseq | XP_019180618.1 | 1e-159 | PREDICTED: MADS-box protein CMB1-like | ||||
| Swissprot | K4BND8 | 1e-103 | MADS4_SOLLC; MADS-box protein 04g005320 | ||||
| TrEMBL | A0A328D358 | 1e-113 | A0A328D358_9ASTE; Uncharacterized protein | ||||
| TrEMBL | A0A484LKQ9 | 1e-113 | A0A484LKQ9_9ASTE; Uncharacterized protein | ||||
| TrEMBL | A0A484M9X4 | 1e-113 | A0A484M9X4_9ASTE; Uncharacterized protein | ||||
| STRING | XP_009788435.1 | 1e-108 | (Nicotiana sylvestris) | ||||
| STRING | PGSC0003DMT400072891 | 1e-108 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA40 | 24 | 625 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G15800.1 | 2e-81 | MIKC_MADS family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



