 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc000131.1_g00007.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 750aa MW: 80987.3 Da PI: 6.5576 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 103.9 | 9e-33 | 287 | 344 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK+vkgse+prsYY+Ct+ +Cpvkkkvers+ +++++ei+Y+g Hnh+k
Itr_sc000131.1_g00007.1 287 DDGYNWRKYGQKQVKGSEYPRSYYKCTHLNCPVKKKVERSQ-EGHITEIIYKGAHNHPK 344
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 32.8 | 1.4e-10 | 500 | 520 | 1 | 21 |
---SS-EEEEEEE--TT-SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefp 21
ldDgy+WrKYGqK+vkg+++p
Itr_sc000131.1_g00007.1 500 LDDGYRWRKYGQKVVKGNPNP 520
59*****************99 PP
| |||||||
| 3 | WRKY | 52.9 | 7.4e-17 | 559 | 598 | 20 | 59 |
S-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 20 fprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+rsYY+CtsagC+v+k+ver+++d k v++tYeg+Hnh+
Itr_sc000131.1_g00007.1 559 STRSYYKCTSAGCTVRKHVERASHDLKSVITTYEGKHNHD 598
68*************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF118290 | 9.68E-26 | 285 | 345 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.2E-28 | 285 | 345 | IPR003657 | WRKY domain |
| SMART | SM00774 | 8.6E-37 | 286 | 344 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.1E-25 | 287 | 343 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.479 | 287 | 345 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.2E-34 | 485 | 520 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.07E-8 | 492 | 521 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 29.666 | 495 | 600 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.3E-29 | 500 | 599 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.3E-6 | 501 | 521 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 5.1E-13 | 554 | 600 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.4E-11 | 556 | 598 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.2E-34 | 561 | 600 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 750 aa Download sequence Send to blast |
MGGFDDHVAI MGDWMPPSPS PRAFFSSILG DDLGTRSAAE PTRENKTGTL VSEPEGYASS 60 RNSDGKGGDQ AGSMSSLSEQ KIGSRGGGLL ERMAARAGFN APRLNTESIR TADLSQNQEV 120 RSPYLTIPPG LSPTTLLDSP VFLSNSLAQP SPTTGKFPFP SSGDIQNSTM FMEASRKSKE 180 NSFDNNDSSS FAFQPVFQTG PSIFPGTFGK VPPSNLARQS FPNVEPTQVY AQNGTIQRPD 240 FSRSSTEKDN GSGNITSEHS PPPDELPDEE TDQRGSGDPN FLGAPADDGY NWRKYGQKQV 300 KGSEYPRSYY KCTHLNCPVK KKVERSQEGH ITEIIYKGAH NHPKPTPNRR SALGSANPLG 360 DMQIENREQA GTGGDGDPIW ANMQKGSGAG DPDWRNDNLE VTTSAPLGSE YCNGSSLQAQ 420 NATQFESGDA VDRSCTFSND EDEDDRGTHG SVSLGYDGEG DESESKRRKI ETYAADMSGA 480 TRAIREPRVV VQTTSEVDIL DDGYRWRKYG QKVVKGNPNP SICSSILSQP VEAQRVNIAS 540 IESRSHDLPY RRVITCHLST RSYYKCTSAG CTVRKHVERA SHDLKSVITT YEGKHNHDVP 600 AARNSSHVNS GVSNSGPSQA TAPNQTHLHR PEPSQLQNTM ARFEGPLSLG SFGLAGRPPQ 660 LGGTPGFGFG MNPQGLTNLA MAGLGANPGK LPVLPVHPYL GQQRPVNHMS FMMSKGEPKM 720 EPVSDPGLNI SNSSSVYQQV MSRLPLGPHM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-29 | 287 | 601 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-29 | 287 | 601 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Regulates WOX8 and WOX9 expression and basal cell division patterns during early embryogenesis. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Required to repolarize the zygote from a transient symmetric state. {ECO:0000269|PubMed:21316593}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
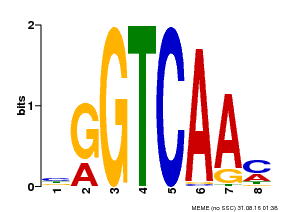
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB026890 | 4e-44 | AB026890.1 Nicotiana tabacum mRNA for transcription factor NtWRKY4, complete cds. | |||
| GenBank | AB063576 | 4e-44 | AB063576.1 Nicotiana tabacum NtWRKY-9 mRNA for WRKY DNA-binding protein, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019196608.1 | 0.0 | PREDICTED: probable WRKY transcription factor 2 isoform X2 | ||||
| Swissprot | Q9FG77 | 0.0 | WRKY2_ARATH; Probable WRKY transcription factor 2 | ||||
| TrEMBL | A0A484M1Z5 | 0.0 | A0A484M1Z5_9ASTE; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400056952 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3711 | 23 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56270.1 | 1e-178 | WRKY DNA-binding protein 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



