 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc000145.1_g00043.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 357aa MW: 38956 Da PI: 7.6201 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 47.7 | 3.3e-15 | 255 | 299 | 5 | 49 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkel 49
+r++r++kNRe+A rsR+RK+a++ eLe+k+ Le+eN +L+ e
Itr_sc000145.1_g00043.1 255 RRQKRMIKNRESAARSRARKQAYTNELENKISRLEEENARLRREK 299
79****************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 2.3E-15 | 250 | 298 | No hit | No description |
| SMART | SM00338 | 2.0E-14 | 251 | 315 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.645 | 253 | 298 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 5.94E-12 | 255 | 309 | No hit | No description |
| CDD | cd14707 | 1.77E-23 | 255 | 297 | No hit | No description |
| Pfam | PF00170 | 3.1E-12 | 255 | 298 | IPR004827 | Basic-leucine zipper domain |
| PROSITE pattern | PS00036 | 0 | 258 | 273 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 357 aa Download sequence Send to blast |
MGSEGGCGGT RMQDPKSNGL EREGSLYSLT LDEVQQQFGG LGKPLSSMNL DELLKTVCTA 60 EAASQGVQQR VDYSGAPLAS GAPLHRQSSL ELSGDLSKKT VDEVWQDIQQ GQKSSSLERK 120 ATLGEMTLED FLVQARIVAE SSQGKNDLGS VQGDDSTVNV AQQAHGALFH VPPIQQPRPP 180 QSHQQQVSAM PGFMAAHPVQ QAIPVGPNPI MDATYPGTQM TAALDPMMGT LSDTQTLGRK 240 RVAPDNVVIE KSVERRQKRM IKNRESAARS RARKQAYTNE LENKISRLEE ENARLRREKN 300 IYSLLIVVLN AMYPGIANLR VSPVLDGKSS EEIEKAMPTV PPPEPKYQLR RTSSAPF |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00409 | DAP | Transfer from AT3G56850 | Download |
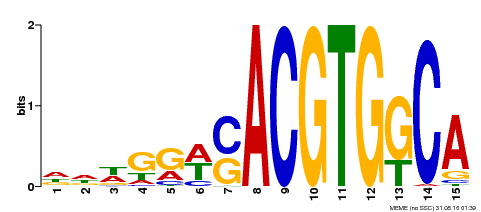
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019186958.1 | 0.0 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 2 isoform X2 | ||||
| TrEMBL | A0A2G3BGU0 | 1e-125 | A0A2G3BGU0_CAPCH; ABSCISIC ACID-INSENSITIVE 5-like protein 2 | ||||
| STRING | Solyc10g081350.1.1 | 1e-116 | (Solanum lycopersicum) | ||||
| STRING | XP_009614068.1 | 1e-116 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3334 | 24 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G56850.1 | 1e-72 | ABA-responsive element binding protein 3 | ||||



