 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc000355.1_g00010.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 551aa MW: 60008.6 Da PI: 6.5237 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 105.8 | 2.2e-33 | 212 | 269 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK+vkgse+prsYY+Ct+++Cp+kkkver+ d++++ei+Y+g Hnh+k
Itr_sc000355.1_g00010.1 212 DDGYNWRKYGQKQVKGSENPRSYYKCTHPNCPTKKKVERAL-DGQITEIVYKGAHNHPK 269
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 105.1 | 3.7e-33 | 387 | 445 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Cts+gCpv+k+ver+++d + v++tYeg+Hnh+
Itr_sc000355.1_g00010.1 387 LDDGYRWRKYGQKVVKGNPNPRSYYKCTSQGCPVRKHVERASHDIRSVITTYEGKHNHD 445
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 3.9E-29 | 202 | 270 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.68E-26 | 206 | 270 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.904 | 206 | 270 | IPR003657 | WRKY domain |
| SMART | SM00774 | 9.5E-37 | 211 | 269 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.0E-25 | 212 | 268 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.6E-37 | 372 | 447 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.27E-29 | 379 | 447 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 39.18 | 382 | 447 | IPR003657 | WRKY domain |
| SMART | SM00774 | 8.5E-39 | 387 | 446 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.5E-25 | 388 | 445 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0010508 | Biological Process | positive regulation of autophagy | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 551 aa Download sequence Send to blast |
MAASSGTIDA PTASSSFSFS TASSFMSSSF TDLLASDAYS GGSVSRGLGD RIAERTGSGV 60 PKFKSLPPPS LPLSSPAVSP SSYFAFPPGL SPSELLDSPV LLSSSNILPS PTTGTFPAQA 120 FNWKNDSNAS QEDVKQEEKG YPDFSFQTNS ASMRTLNYED SKRKDELNSL QSLPPVTTST 180 QMSSQNNGGS YSEYNNQCCP PSQTLREQRR SDDGYNWRKY GQKQVKGSEN PRSYYKCTHP 240 NCPTKKKVER ALDGQITEIV YKGAHNHPKP QSTRRSSSST ASSASTLAAQ SYNPPASDVP 300 DQSYWSNGNG QMDSVATPEN SSISVGDDEF EQSSQKREPV GDEFDEDEPD AKRWKVENES 360 EGVSAQGSRT VREPRVVVQT TSDIDILDDG YRWRKYGQKV VKGNPNPRSY YKCTSQGCPV 420 RKHVERASHD IRSVITTYEG KHNHDVPAAR GSGSHGLNRG ANPNNNAAMA MAIRPSTMSL 480 QSNYPIPIPS TRAMQPGEGQ APYEMLQGPG GFGYSGFGNP MNAYANQIQD NAFSRAKEEP 540 RDDLFLERLL A |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 5e-36 | 203 | 448 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 5e-36 | 203 | 448 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:26025535). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (PubMed:19199048, PubMed:26025535). Negative regulator of both gibberellic acid (GA) and abscisic acid (ABA) signaling in aleurone cells, probably by interfering with GAM1, via the specific repression of GA- and ABA-induced promoters (PubMed:15618416, PubMed:19199048, PubMed:26025535). {ECO:0000269|PubMed:15618416, ECO:0000269|PubMed:19199048, ECO:0000269|PubMed:26025535}. | |||||
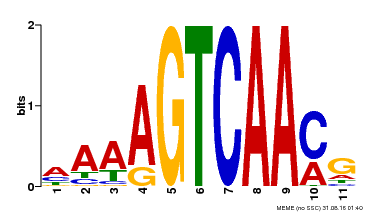
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00299 | DAP | Transfer from AT2G38470 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) in aleurone cells, embryos, roots and leaves (PubMed:15618416, PubMed:19199048). Slightly down-regulated by gibberellic acid (GA) (PubMed:15618416). Accumulates in response to jasmonic acid (MeJA) (By similarity). {ECO:0000250|UniProtKB:Q6B6R4, ECO:0000269|PubMed:15618416, ECO:0000269|PubMed:19199048}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KU323590 | 0.0 | KU323590.1 Ipomoea trifida WRKY protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019178621.1 | 0.0 | PREDICTED: probable WRKY transcription factor 26 isoform X1 | ||||
| Swissprot | Q6IEQ7 | 1e-137 | WRK24_ORYSJ; WRKY transcription factor WRKY24 | ||||
| TrEMBL | A0A191T7X3 | 0.0 | A0A191T7X3_IPOTF; WRKY protein | ||||
| STRING | EOY34631 | 0.0 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2286 | 24 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38470.1 | 1e-116 | WRKY DNA-binding protein 33 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



