 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc000417.1_g00057.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 336aa MW: 36683.9 Da PI: 8.8632 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 130.3 | 6.7e-41 | 26 | 102 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
C v+gC+adls+++eyhrrhkvCe hsk+p+v++ g+eqrfCqqCsrfh+l efDe krsCr+rL++hn+rrrk+q+
Itr_sc000417.1_g00057.1 26 CLVDGCNADLSRCREYHRRHKVCELHSKTPKVTIGGREQRFCQQCSRFHSLAEFDEVKRSCRKRLDGHNRRRRKPQP 102
**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.7E-33 | 20 | 87 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.983 | 23 | 100 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.01E-38 | 24 | 105 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.6E-30 | 26 | 99 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 336 aa Download sequence Send to blast |
MESSSSLSGS SKRAKAPGSV GQVAHCLVDG CNADLSRCRE YHRRHKVCEL HSKTPKVTIG 60 GREQRFCQQC SRFHSLAEFD EVKRSCRKRL DGHNRRRRKP QPESLSKSSG MLFSGQQGAK 120 LLSFSSSQIF PTSSVSSTWP GFSKPENDVV LYSNNQSQSH LNYIIDTQNS FPDSCAQFKG 180 VMNQFQFMQG SSEASSISQQ QQPLVDHNSA RGNCSQKLFA AAGLSSRIVD SDRALSLLSS 240 ASSSSVNREI GLSHVVQPEA IPHPLHYGGL ISHYPFVAHE IETKPAGVSA AAAADCHAIK 300 NAARLQFHNV IPNAPTTDVS SSGGPHQTHT FMWEQD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 7e-28 | 26 | 99 | 11 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 82 | 99 | KRSCRKRLDGHNRRRRKP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
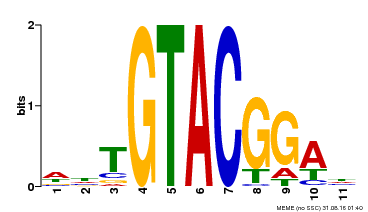
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00555 | DAP | Transfer from AT5G50570 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156 and miR157. {ECO:0000305|PubMed:12202040}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GU172038 | 1e-98 | GU172038.1 Ipomoea batatas microsatellite Stv_Ipb_1841 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019180911.1 | 1e-165 | PREDICTED: squamosa promoter-binding-like protein 13A isoform X2 | ||||
| Swissprot | B9DI20 | 7e-51 | SP13A_ARATH; Squamosa promoter-binding-like protein 13A | ||||
| Swissprot | P0DI11 | 7e-51 | SP13B_ARATH; Squamosa promoter-binding-like protein 13B | ||||
| TrEMBL | A0A125SZN9 | 1e-123 | A0A125SZN9_TOBAC; Squamosa promoter binding protein NtabSPL13 | ||||
| TrEMBL | A0A314KI71 | 1e-123 | A0A314KI71_NICAT; Squamosa promoter-binding-like protein 16 | ||||
| STRING | XP_009621034.1 | 1e-124 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4920 | 20 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G50670.1 | 3e-47 | SBP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



