 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc000499.1_g00002.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 396aa MW: 44511.6 Da PI: 6.7707 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 28.5 | 2.7e-09 | 324 | 358 | 21 | 55 |
HSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 21 knrypsaeereeLAkklgLterqVkvWFqNrRake 55
k +yps++e+ LA+ +gL+++q+ +WF N+R ++
Itr_sc000499.1_g00002.1 324 KWPYPSETEKVALAEATGLDQKQINNWFINQRKRH 358
669*****************************885 PP
| |||||||
| 2 | ELK | 35.9 | 1.6e-12 | 278 | 299 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELK++Ll+KYsgyL+sLkqE+s
Itr_sc000499.1_g00002.1 278 ELKNHLLKKYSGYLSSLKQELS 299
9*******************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 1.9E-22 | 120 | 164 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 3.6E-23 | 121 | 163 | IPR005540 | KNOX1 |
| SMART | SM01256 | 3.8E-27 | 172 | 223 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 6.1E-23 | 177 | 221 | IPR005541 | KNOX2 |
| PROSITE profile | PS51213 | 11.243 | 278 | 298 | IPR005539 | ELK domain |
| SMART | SM01188 | 1.0E-6 | 278 | 299 | IPR005539 | ELK domain |
| Pfam | PF03789 | 7.9E-10 | 278 | 299 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.811 | 298 | 361 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 2.05E-19 | 299 | 372 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 1.5E-12 | 300 | 365 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 4.0E-27 | 303 | 364 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 3.93E-12 | 310 | 362 | No hit | No description |
| Pfam | PF05920 | 1.7E-16 | 318 | 357 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 336 | 359 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001708 | Biological Process | cell fate specification | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 396 aa Download sequence Send to blast |
MENYGQLGEE TAAAAARQRG SGTGFMYGSS VLGPPSYGRT GGESNHVATG FHLQPGGGGE 60 CYGQSDGPPH TTVKSEGGGS SASHHHHHHH QMFQYPLMIR DHLHPRQQES AGDNSLSEGD 120 AMKAKIIAHP QYSNLLEAYV DCQKVGAPPE MAARLSAARQ ELEGKQRASF IGSRDSSKDP 180 ELDQFMEAYY DMLMKYRDEL TRPFQEAMEF MRRIESQLNM LSDGPVRIFN SGGVKGSNAR 240 EHLVDDKCEG VGSSEEDQDN SGGETELREI DPRVQDRELK NHLLKKYSGY LSSLKQELSK 300 KKKKGKLPKD ARQKLLSWWE LHYKWPYPSE TEKVALAEAT GLDQKQINNW FINQRKRHWK 360 PSEDMQFMVM DGLHHQNAAL YMDGHYMGDA PYRLGP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Appears to be involved in meristem formation and in the regulation of leaf morphology. Misexpression makes the leaf more compound which is always associated with growth retardation and loss of apical dominance, resulting in dwarfed, bushy plants. Probably binds to the DNA sequence 5'-TGAC-3'. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00675 | ChIP-seq | Transfer from GRMZM2G017087 | Download |
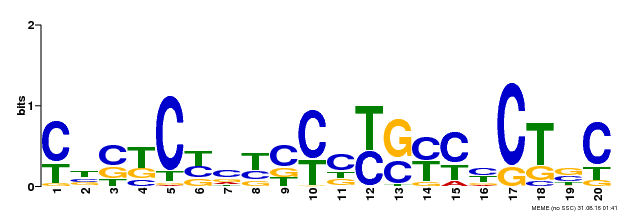
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB283028 | 0.0 | AB283028.1 Ipomoea batatas Ibkn2 mRNA for class-I knotted1-like homeobox protein IBKN2, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019174076.1 | 0.0 | PREDICTED: homeotic protein knotted-1-like | ||||
| Swissprot | Q41330 | 1e-166 | KN1_SOLLC; Homeotic protein knotted-1 | ||||
| TrEMBL | A8QXP6 | 0.0 | A8QXP6_IPOBA; Class-I knotted1-like homeobox protein IBKN2 | ||||
| STRING | EOY15624 | 1e-180 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4973 | 23 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 1e-129 | KNOTTED-like from Arabidopsis thaliana | ||||



